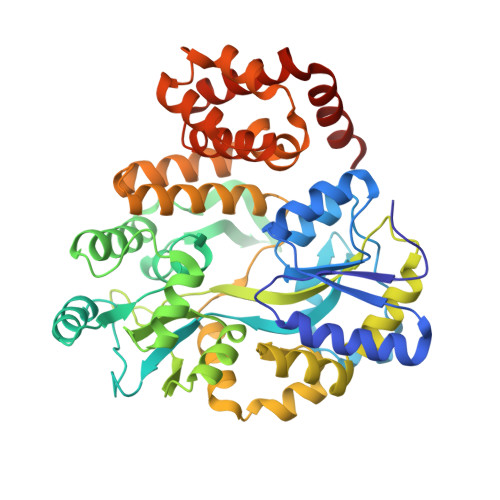
Structure of the NLRP1 caspase recruitment domain suggests potential mechanisms for its association with procaspase-1.
Jin, T., Curry, J., Smith, P., Jiang, J., Xiao, T.S.(2013) Proteins 81: 1266-1270
- PubMed: 23508996
- DOI: https://doi.org/10.1002/prot.24287
- Primary Citation of Related Structures:
4IFP - PubMed Abstract:
The NLRP1 inflammasome responds to microbial challenges such as Bacillus anthracis infection and is implicated in autoimmune disease such as vitiligo. Human NLRP1 contains both an N-terminal pyrin domain (PYD) and a C-terminal caspase recruitment domain (CARD), with the latter being essential for its association with the downstream effector procaspase-1. Here we report a 2.0 Å crystal structure of the human NLRP1 CARD as a fusion with the maltose-binding protein. The structure reveals the six-helix bundle fold of the NLRP1 CARD, typical of the death domain superfamily. The charge surface of the NLRP1 CARD structure and a procaspase-1 CARD model suggests potential mechanisms for their association through electrostatic attraction.
- Structural Immunobiology Unit, Laboratory of Immunology, National Institute of Allergy and Infectious Diseases, National Institutes of Health, Bethesda, Maryland 20892-0430, USA.
Organizational Affiliation: