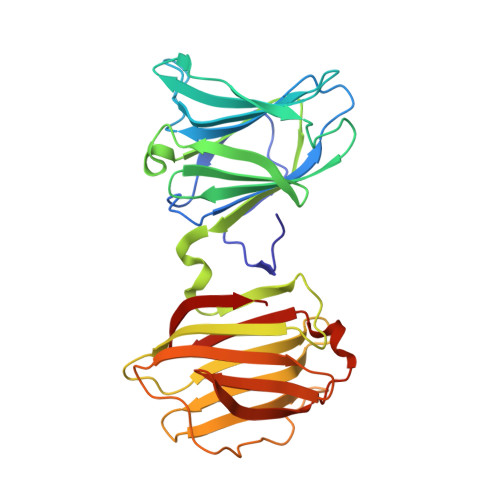
X-ray structure of a protease-resistant mutant form of human galectin-8 with two carbohydrate recognition domains
Yoshida, H., Yamashita, S., Teraoka, M., Itoh, A., Nakakita, S., Nishi, N., Kamitori, S.(2012) FEBS J 279: 3937-3951
- PubMed: 22913484
- DOI: https://doi.org/10.1111/j.1742-4658.2012.08753.x
- Primary Citation of Related Structures:
3VKL, 3VKM, 3VKN, 3VKO, 4FQZ - PubMed Abstract:
Galectin-8 is a tandem-repeat-type β-galactoside-specific animal lectin possessing N-terminal and C-terminal carbohydrate recognition domains (N-CRD and C-CRD, respectively), with a difference in carbohydrate-binding specificity, involved in cell-matrix interaction, malignant transformation, and cell adhesion. N-CRD shows strong affinity for α2-3-sialylated oligosaccharides, a feature unique to galectin-8. C-CRD usually shows lower affinity for oligosaccharides but higher affinity for N-glycan-type branched oligosaccharides than does N-CRD. There have been many structural studies on galectins with a single carbohydrate recognition domain (CRD), but no X-ray structure of a galectin containing both CRDs has been reported. Here, the X-ray structure of a protease-resistant mutant form of human galectin-8 possessing both CRDs and the novel pseudodimer structure of galectin-8 N-CRD in complexes with α2-3-sialylated oligosaccharide ligands were determined. The results revealed a difference in specificity between N-CRD and C-CRD, and provided new insights into the association of CRDs and/or molecules of galectin-8.
- Life Science Research Center and Faculty of Medicine, Kagawa University, Kagawa, Japan.
Organizational Affiliation: