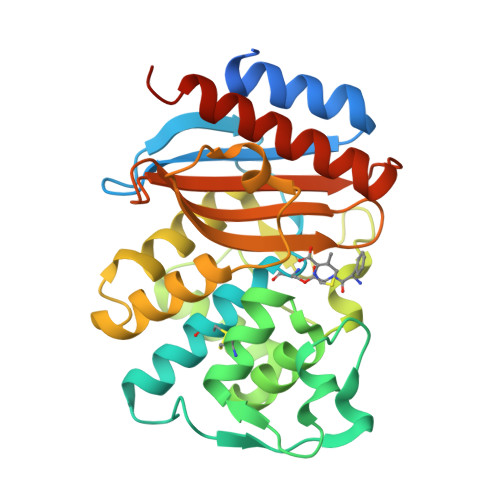
Exploring the potential impact of an expanded genetic code on protein function.
Xiao, H., Nasertorabi, F., Choi, S.H., Han, G.W., Reed, S.A., Stevens, R.C., Schultz, P.G.(2015) Proc Natl Acad Sci U S A 112: 6961-6966
- PubMed: 26038548
- DOI: https://doi.org/10.1073/pnas.1507741112
- Primary Citation of Related Structures:
4ZJ1, 4ZJ2, 4ZJ3 - PubMed Abstract:
With few exceptions, all living organisms encode the same 20 canonical amino acids; however, it remains an open question whether organisms with additional amino acids beyond the common 20 might have an evolutionary advantage. Here, we begin to test that notion by making a large library of mutant enzymes in which 10 structurally distinct noncanonical amino acids were substituted at single sites randomly throughout TEM-1 β-lactamase. A screen for growth on the β-lactam antibiotic cephalexin afforded a unique p-acrylamido-phenylalanine (AcrF) mutation at Val-216 that leads to an increase in catalytic efficiency by increasing kcat, but not significantly affecting KM. To understand the structural basis for this enhanced activity, we solved the X-ray crystal structures of the ligand-free mutant enzyme and of the deacylation-defective wild-type and mutant cephalexin acyl-enzyme intermediates. These structures show that the Val-216-AcrF mutation leads to conformational changes in key active site residues-both in the free enzyme and upon formation of the acyl-enzyme intermediate-that lower the free energy of activation of the substrate transacylation reaction. The functional changes induced by this mutation could not be reproduced by substitution of any of the 20 canonical amino acids for Val-216, indicating that an expanded genetic code may offer novel solutions to proteins as they evolve new activities.
Organizational Affiliation:
Department of Chemistry and the Skaggs Institute for Chemical Biology, The Scripps Research Institute, La Jolla, CA 92037;