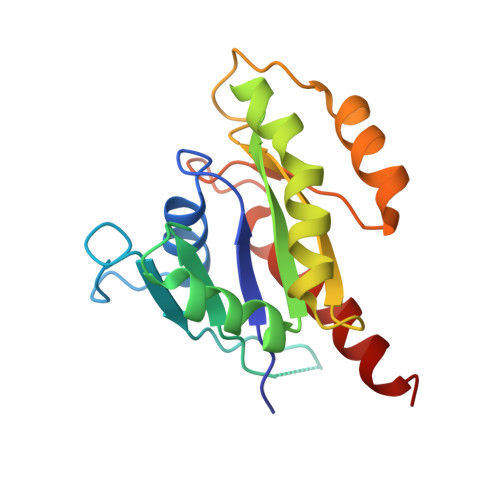
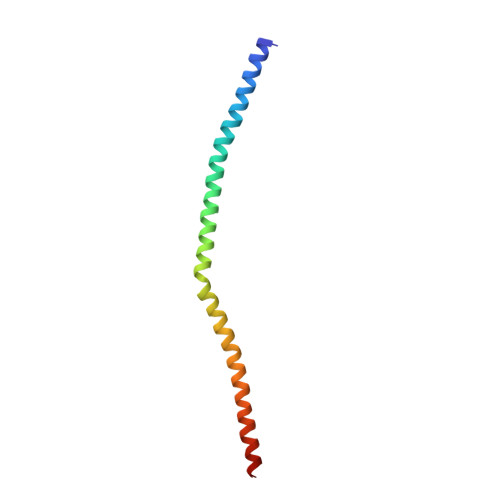New insights into the molecular mechanism of the Rab GTPase Sec4p activation.
Rinaldi, F.C., Packer, M., Collins, R.(2015) BMC Struct Biol 15: 14-14
- PubMed: 26263895
- DOI: https://doi.org/10.1186/s12900-015-0041-5
- Primary Citation of Related Structures:
4Z8Y, 4ZDW - PubMed Abstract:
Sec4p is a small monomeric Ras-related GTP-binding protein (23 kDa) that regulates polarized exocytosis in S. cerevisiae. In this study we examine the structural effects of a conserved serine residue in the P-loop corresponding to G12 in Ras. We show that the Sec4p residue serine 29 forms a hydrogen bond with the nucleotide. Mutations of this residue have a different impact than equivalent mutations in Ras and can form stable associations with the exchange factor allowing us to elucidate the structure of a complex of Sec4p bound to the exchange factor Sec2p representing an early stage of the exchange reaction. Our structural investigation of the Sec4p-Sec2p complex reveals the role of the Sec2p coiled-coil domain in facilitating the fast kinetics of the exchange reaction. For Ras-family GTPases, single point mutations that impact the signaling state of the molecule have been well described however less structural information is available for equivalent mutations in the case of Rab proteins. Understanding the structural properties of mutants such as the one described here, provides useful insights into unique aspects of Rab GTPase function.
Organizational Affiliation:
Department of Molecular Medicine, Cornell University, Ithaca, NY, 14853, USA. fcr23@cornell.edu.