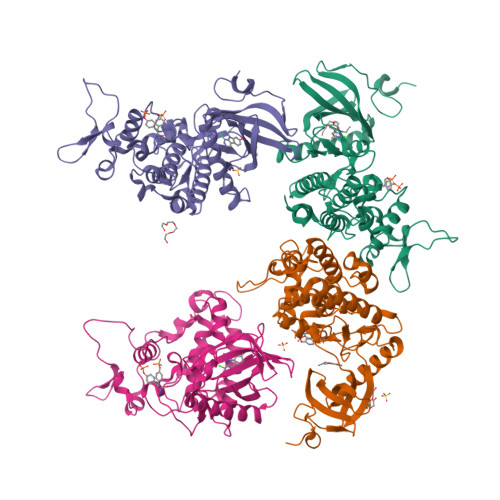
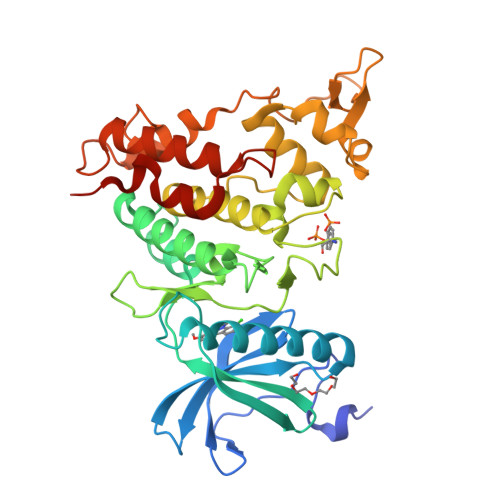
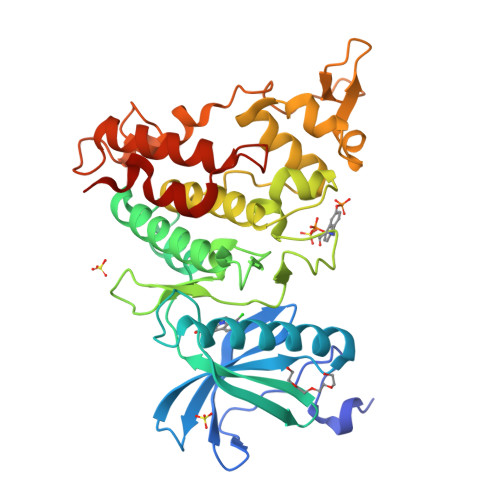
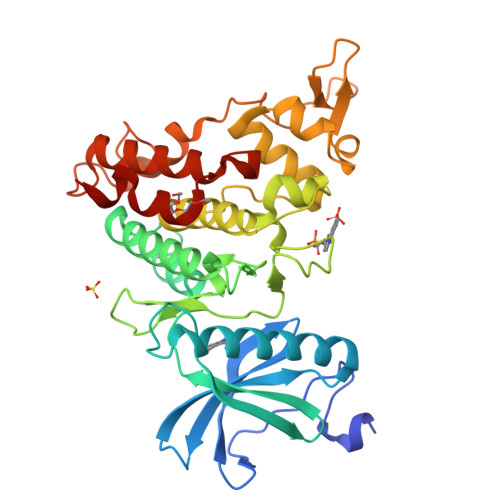
How to Separate Kinase Inhibition from Undesired Monoamine Oxidase A Inhibition-The Development of the DYRK1A Inhibitor AnnH75 from the Alkaloid Harmine.
Wurzlbauer, A., Ruben, K., Gurdal, E., Chaikuad, A., Knapp, S., Sippl, W., Becker, W., Bracher, F.(2020) Molecules 25
- PubMed: 33339338
- DOI: https://doi.org/10.3390/molecules25245962
- Primary Citation of Related Structures:
4YU2 - PubMed Abstract:
The β-carboline alkaloid harmine is a potent DYRK1A inhibitor, but suffers from undesired potent inhibition of MAO-A, which strongly limits its application. We synthesized more than 60 analogues of harmine, either by direct modification of the alkaloid or by de novo synthesis of β-carboline and related scaffolds aimed at learning about structure-activity relationships for inhibition of both DYRK1A and MAO-A, with the ultimate goal of separating desired DYRK1A inhibition from undesired MAO-A inhibition. Based on evidence from published crystal structures of harmine bound to each of these enzymes, we performed systematic structure modifications of harmine yielding DYRK1A-selective inhibitors characterized by small polar substituents at N-9 (which preserve DYRK1A inhibition and eliminate MAO-A inhibition) and beneficial residues at C-1 (methyl or chlorine). The top compound AnnH75 remains a potent DYRK1A inhibitor, and it is devoid of MAO-A inhibition. Its binding mode to DYRK1A was elucidated by crystal structure analysis, and docking experiments provided additional insights for this attractive series of DYRK1A and MAO-A inhibitors.
Organizational Affiliation:
Department of Pharmacy-Center for Drug Research, Ludwig-Maximilians University, 81377 Munich, Germany.