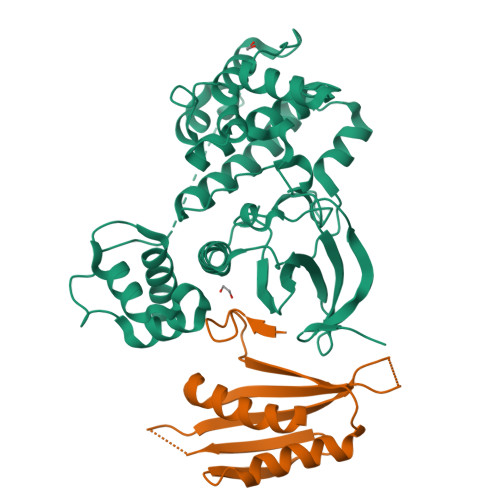
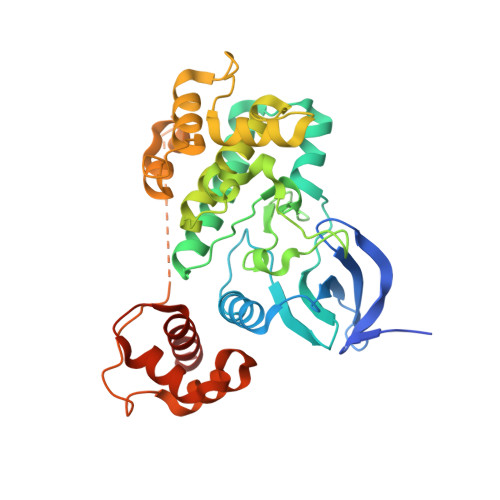
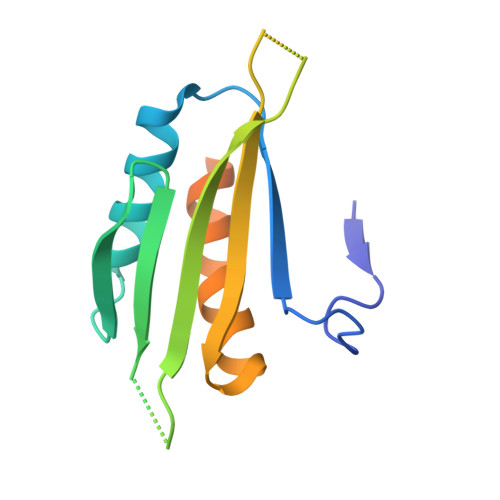
Structural insight into the mechanism of synergistic autoinhibition of SAD kinases
Wu, J.X., Cheng, Y.S., Wang, J., Chen, L., Ding, M., Wu, J.W.(2015) Nat Commun 6: 8953-8953
- PubMed: 26626945
- DOI: https://doi.org/10.1038/ncomms9953
- Primary Citation of Related Structures:
4YNZ, 4YOM - PubMed Abstract:
The SAD/BRSK kinases participate in various important life processes, including neural development, cell cycle and energy metabolism. Like other members of the AMPK family, SAD contains an N-terminal kinase domain followed by the characteristic UBA and KA1 domains. Here we identify a unique autoinhibitory sequence (AIS) in SAD kinases, which exerts autoregulation in cooperation with UBA. Structural studies of mouse SAD-A revealed that UBA binds to the kinase domain in a distinct mode and, more importantly, AIS nestles specifically into the KD-UBA junction. The cooperative action of AIS and UBA results in an 'αC-out' inactive kinase, which is conserved across species and essential for presynaptic vesicle clustering in C. elegans. In addition, the AIS, along with the KA1 domain, is indispensable for phospholipid binding. Taken together, these data suggest a model for synergistic autoinhibition and membrane activation of SAD kinases.
Organizational Affiliation:
MOE Key Laboratory for Protein Science and Tsinghua-Peking Center for Life Sciences, School of Life Sciences, Tsinghua University, Beijing 100084, China.