Discovery of 1,3-dihydro-2,1,3-benzothiadiazole 2,2-dioxide analogs as new RORC modulators.
Muegge, I., Collin, D., Cook, B., Hill-Drzewi, M., Horan, J., Kugler, S., Labadia, M., Li, X., Smith, L., Zhang, Y.(2015) Bioorg Med Chem Lett 25: 1892-1895
- PubMed: 25840886
- DOI: https://doi.org/10.1016/j.bmcl.2015.03.042
- Primary Citation of Related Structures:
4YMQ - PubMed Abstract:
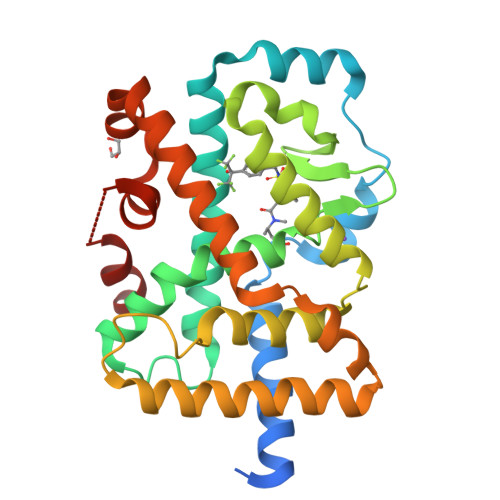
Structure-based and pharmacophore-based virtual screening in combination with combinatorial chemistry and X-ray crystallography led to the discovery of a new class of benzothiadiazole dioxide analogs with functional activity as RORC inverse agonists. The early RORC SAR compound 14 exhibited RORC inhibition in a cell based reporter gene assay of 5.7 μM and bound to RORC with an affinity of 1.6 μM in a fluorescence polarization assay displacing a ligand binding site probe. Crystallography confirmed the binding mode of the compound in the ligand binding domain displaying the engagement of a novel sub pocket close to Ser404. Subsequent optimization yielded compounds with enhanced RORC inverse agonist activity. The most active compound 19 showed an IC50 of 440 nM in a human PBMC assay.
Organizational Affiliation:
Boehringer Ingelheim Pharmaceuticals, Inc., 900 Ridgebury Road, PO Box 368, Ridgefield, CT 06877-0368, USA. Electronic address: ingo.mugge@boehringer-ingelheim.com.