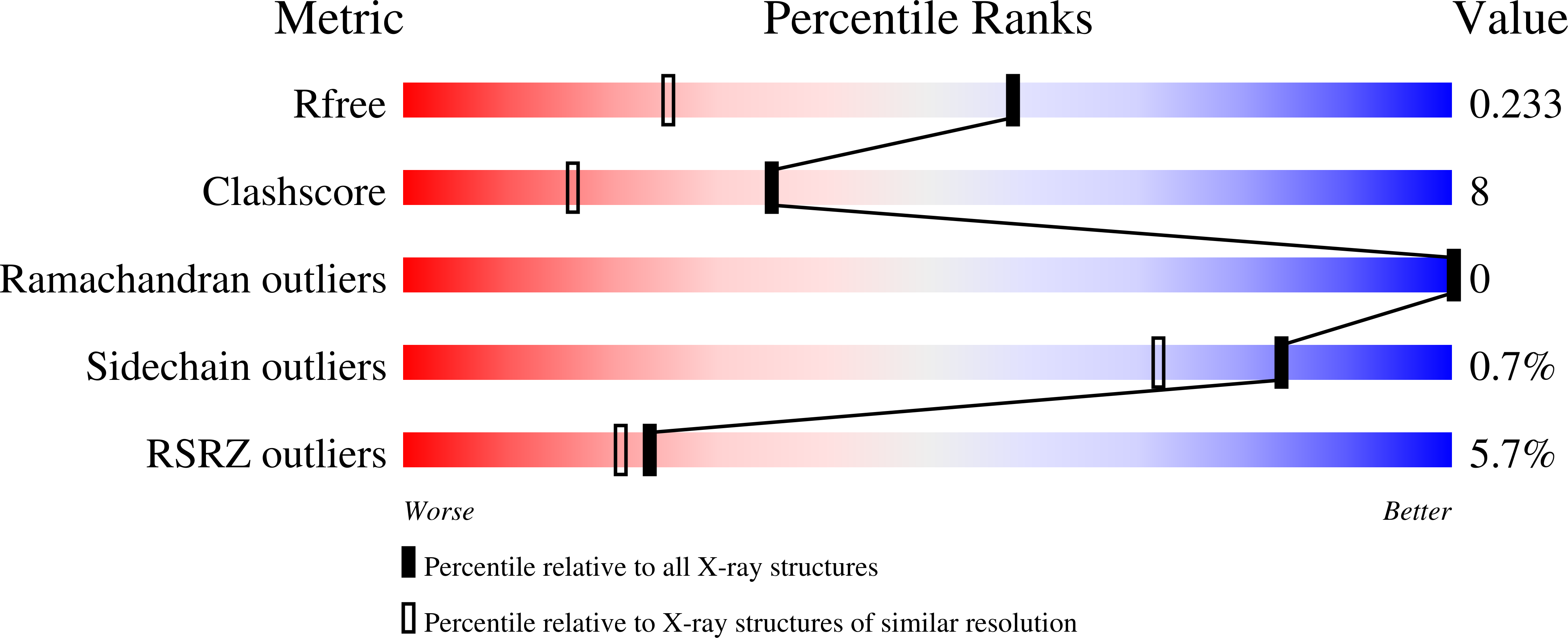
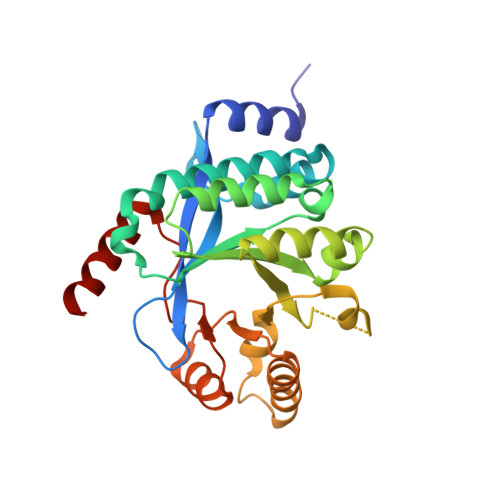
Structure of PA3825 from P. aeruginosa bound to cyclic di-GMP and pGpG: new insights for a potential three-metal catalytic mechanism of EAL domains
Bellini, D., Horrell, S., Strange, R., Wagner, A., Hitchin, A., Webb, J.S., Tews, I., Walch, M.A.To be published.