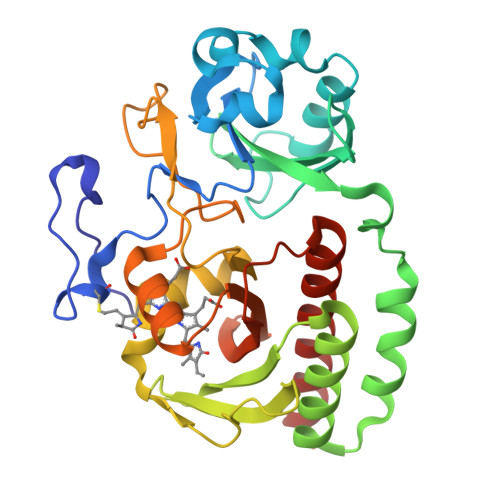
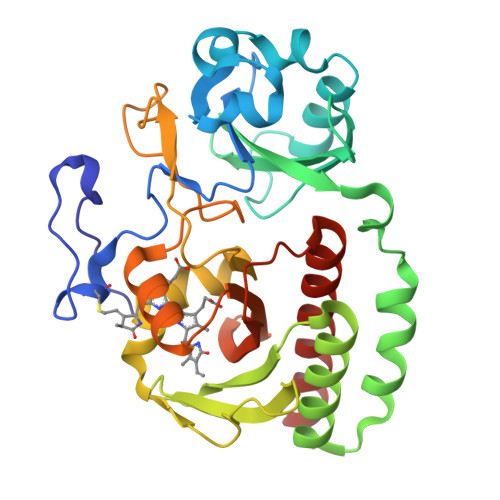
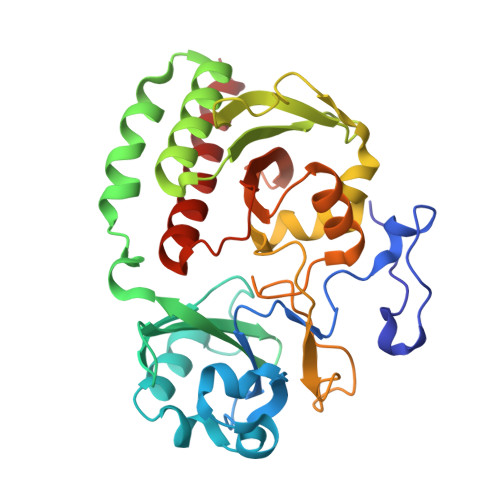
X-ray radiation induces deprotonation of the bilin chromophore in crystalline D. radiodurans phytochrome.
Li, F., Burgie, E.S., Yu, T., Heroux, A., Schatz, G.C., Vierstra, R.D., Orville, A.M.(2015) J Am Chem Soc 137: 2792-2795
- PubMed: 25650486
- DOI: https://doi.org/10.1021/ja510923m
- Primary Citation of Related Structures:
4Y3I, 4Y5F - PubMed Abstract:
We report that in the red light-absorbing (Pr) state, the bilin chromophore of the Deinococcus radiodurans proteobacterial phytochrome (DrBphP) is hypersensitive to X-ray photons used in typical synchrotron X-ray protein crystallography experiments. This causes the otherwise fully protonated chromophore to deprotonate without additional major structural changes. These results have major implications for our understanding of the structural and chemical characteristics of the resting and intermediate states of phytochromes and other photoreceptor proteins.
Organizational Affiliation:
Photon Sciences Directorate and ∥Biosciences Department, Brookhaven National Laboratory , Upton, New York 11973, United States.