Structural Basis for Translocation of a Biofilm-supporting Exopolysaccharide across the Bacterial Outer Membrane
Wang, Y., Andole Pannuri, A., Ni, D., Zhou, H., Cao, X., Lu, X., Romeo, T., Huang, Y.(2016) J Biological Chem 291: 10046-10057
- PubMed: 26957546
- DOI: https://doi.org/10.1074/jbc.M115.711762
- Primary Citation of Related Structures:
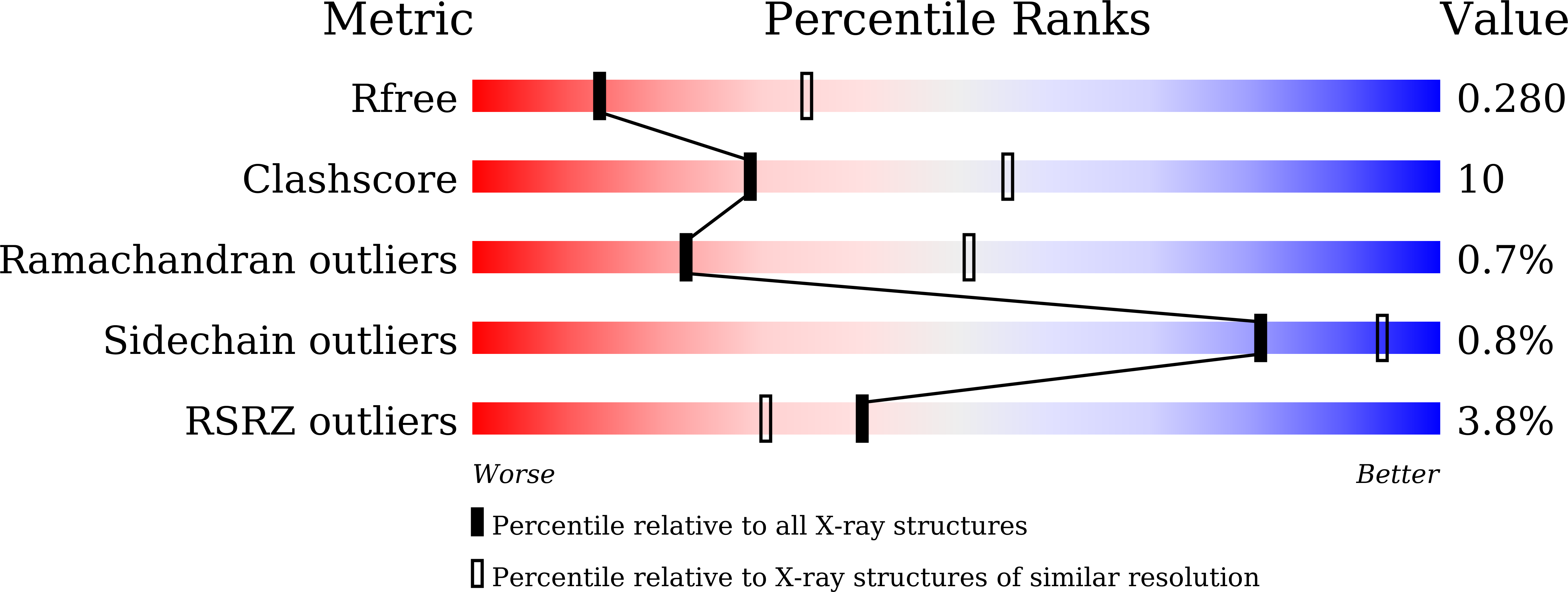
4Y25 - PubMed Abstract:
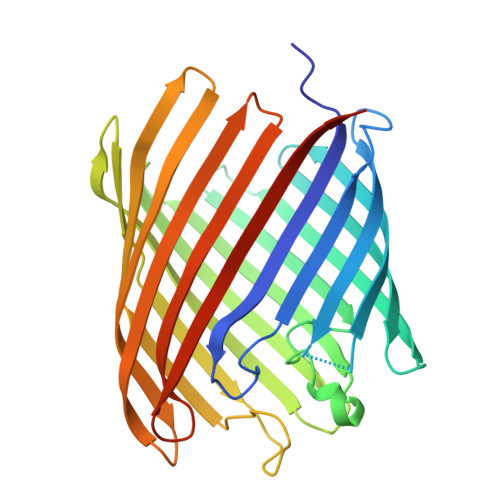
The partially de-N-acetylated poly-β-1,6-N-acetyl-d-glucosamine (dPNAG) polymer serves as an intercellular biofilm adhesin that plays an essential role for the development and maintenance of integrity of biofilms of diverse bacterial species. Translocation of dPNAG across the bacterial outer membrane is mediated by a tetratricopeptide repeat-containing outer membrane protein, PgaA. To understand the molecular basis of dPNAG translocation, we determined the crystal structure of the C-terminal transmembrane domain of PgaA (residues 513-807). The structure reveals that PgaA forms a 16-strand transmembrane β-barrel, closed by four loops on the extracellular surface. Half of the interior surface of the barrel that lies parallel to the translocation pathway is electronegative, suggesting that the corresponding negatively charged residues may assist the secretion of the positively charged dPNAG polymer. In vivo complementation assays in a pgaA deletion bacterial strain showed that a cluster of negatively charged residues proximal to the periplasm is necessary for biofilm formation. Biochemical analyses further revealed that the tetratricopeptide repeat domain of PgaA binds directly to the N-deacetylase PgaB and is critical for biofilm formation. Our studies support a model in which the positively charged PgaB-bound dPNAG polymer is delivered to PgaA through the PgaA-PgaB interaction and is further targeted to the β-barrel lumen of PgaA potentially via a charge complementarity mechanism, thus priming the translocation of dPNAG across the bacterial outer membrane.
Organizational Affiliation:
From the National Laboratory of Biomacromolecules, Institute of Biophysics, Chinese Academy of Sciences, Beijing 100101, China, University of Chinese Academy of Sciences, Beijing 100049, China.