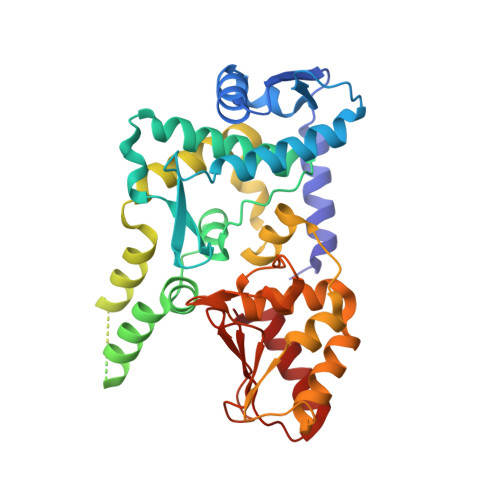
Structure of the HECT domain of human WWP2
Gong, W., Zhang, X., Zhang, W., Li, J., Li, Z.(2015) Acta Crystallogr F Struct Biol Commun 71: 1251-1257
- PubMed: 26457515
- DOI: https://doi.org/10.1107/S2053230X1501554X
- Primary Citation of Related Structures:
4Y07 - PubMed Abstract:
WWP2 is a HECT-domain ubiquitin ligase of the Nedd4 family, which is involved in various important biological processes, such as protein degradation, membrane-protein sorting and transportation, the immune response, pluripotency of embryonic stem cells, tumourigenesis and metastasis. The HECT domain provides the intrinsic ubiquitin ligase activity of WWP2. Here, the expression, purification, crystallization and crystallographic analysis of the HECT domain of human WWP2 (HECT(WWP2)) are reported. HECT(WWP2) has been crystallized and the crystals diffracted to 2.50 Å resolution. They belonged to space group P41212 and the structure has been solved via molecular replacement. The overall structure of HECT(WWP2) has an inverted T-shape. This structure displays a high degree of conservation with previously published structures of Nedd4 subfamily members.
Organizational Affiliation:
Institutes of Biomedical Sciences and School of Basic Medical Sciences, Fudan University, People's Republic of China.