Crystal structure of yeast N-terminal acetyltransferase NatE (ppGpp) in complex with a bisubstrate
Dong, J., Wang, S., York, J.D.To be published.
Experimental Data Snapshot
Entity ID: 1 | |||||
|---|---|---|---|---|---|
| Molecule | Chains | Sequence Length | Organism | Details | Image |
| N-terminal acetyltransferase A complex subunit NAT1 | 854 | Saccharomyces cerevisiae | Mutation(s): 0 | 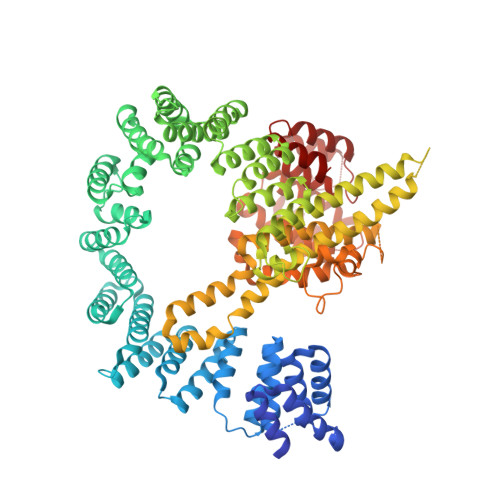 | |
UniProt | |||||
Find proteins for P12945 (Saccharomyces cerevisiae (strain ATCC 204508 / S288c)) Explore P12945 Go to UniProtKB: P12945 | |||||
Entity Groups | |||||
| Sequence Clusters | 30% Identity50% Identity70% Identity90% Identity95% Identity100% Identity | ||||
| UniProt Group | P12945 | ||||
Sequence AnnotationsExpand | |||||
| |||||
Entity ID: 2 | |||||
|---|---|---|---|---|---|
| Molecule | Chains | Sequence Length | Organism | Details | Image |
| N-terminal acetyltransferase A complex catalytic subunit ARD1 | 238 | Saccharomyces cerevisiae | Mutation(s): 0 EC: 2.3.1.88 (PDB Primary Data), 2.3.1.255 (UniProt) | 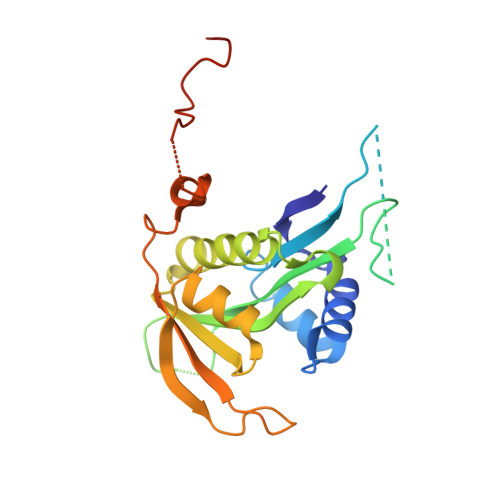 | |
UniProt | |||||
Find proteins for P07347 (Saccharomyces cerevisiae (strain ATCC 204508 / S288c)) Explore P07347 Go to UniProtKB: P07347 | |||||
Entity Groups | |||||
| Sequence Clusters | 30% Identity50% Identity70% Identity90% Identity95% Identity100% Identity | ||||
| UniProt Group | P07347 | ||||
Sequence AnnotationsExpand | |||||
| |||||
Entity ID: 3 | |||||
|---|---|---|---|---|---|
| Molecule | Chains | Sequence Length | Organism | Details | Image |
| N-terminal acetyltransferase A complex subunit NAT5 | 176 | Saccharomyces cerevisiae | Mutation(s): 0 EC: 2.3.1 (PDB Primary Data), 2.3.1.258 (UniProt) | 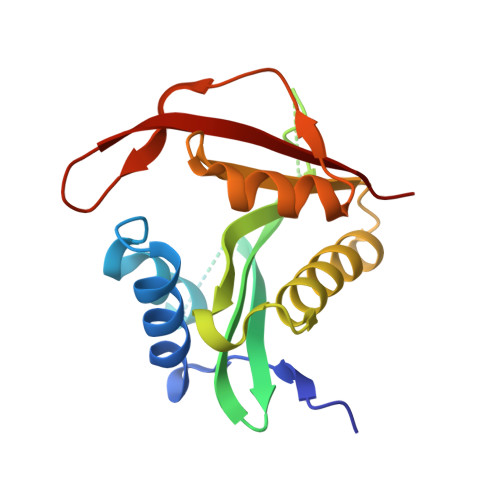 | |
UniProt | |||||
Find proteins for Q08689 (Saccharomyces cerevisiae (strain ATCC 204508 / S288c)) Explore Q08689 Go to UniProtKB: Q08689 | |||||
Entity Groups | |||||
| Sequence Clusters | 30% Identity50% Identity70% Identity90% Identity95% Identity100% Identity | ||||
| UniProt Group | Q08689 | ||||
Sequence AnnotationsExpand | |||||
| |||||
Find similar proteins by: Sequence | 3D Structure
Entity ID: 4 | |||||
|---|---|---|---|---|---|
| Molecule | Chains | Sequence Length | Organism | Details | Image |
| human ACTH8 | D [auth F] | 8 | Homo sapiens | Mutation(s): 0 |  |
UniProt & NIH Common Fund Data Resources | |||||
Find proteins for P01189 (Homo sapiens) Explore P01189 Go to UniProtKB: P01189 | |||||
PHAROS: P01189 GTEx: ENSG00000115138 | |||||
Entity Groups | |||||
| UniProt Group | P01189 | ||||
Sequence AnnotationsExpand | |||||
| |||||
| Ligands 3 Unique | |||||
|---|---|---|---|---|---|
| ID | Chains | Name / Formula / InChI Key | 2D Diagram | 3D Interactions | |
| CMC Query on CMC | G [auth F] | CARBOXYMETHYL COENZYME *A C23 H38 N7 O18 P3 S OBUOSIHPWVNVJN-GRFIIANRSA-N |  | ||
| ACO Query on ACO | F [auth C] | ACETYL COENZYME *A C23 H38 N7 O17 P3 S ZSLZBFCDCINBPY-ZSJPKINUSA-N |  | ||
| G4P Query on G4P | E [auth A] | GUANOSINE-5',3'-TETRAPHOSPHATE C10 H17 N5 O17 P4 BUFLLCUFNHESEH-UUOKFMHZSA-N |  | ||
| Length ( Å ) | Angle ( ˚ ) |
|---|---|
| a = 85.564 | α = 90 |
| b = 113.649 | β = 90 |
| c = 146.721 | γ = 90 |
| Software Name | Purpose |
|---|---|
| REFMAC | refinement |
| HKL-2000 | data reduction |
| HKL-2000 | data scaling |
| PHASER | phasing |