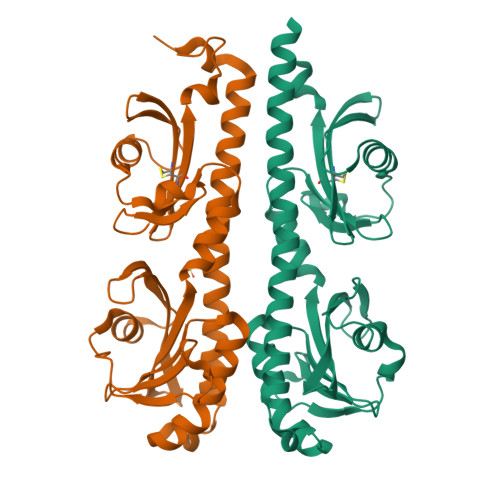
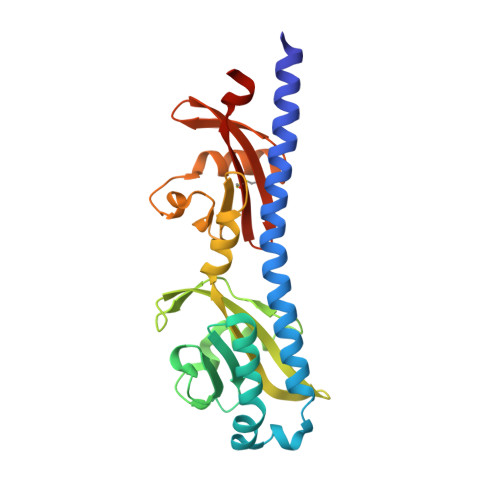
Structural basis for amino-acid recognition and transmembrane signalling by tandem Per-Arnt-Sim (tandem PAS) chemoreceptor sensory domains.
Liu, Y.C., Machuca, M.A., Beckham, S.A., Gunzburg, M.J., Roujeinikova, A.(2015) Acta Crystallogr D Biol Crystallogr 71: 2127-2136
- PubMed: 26457436
- DOI: https://doi.org/10.1107/S139900471501384X
- Primary Citation of Related Structures:
4XMQ, 4XMR - PubMed Abstract:
Chemotaxis, mediated by methyl-accepting chemotaxis protein (MCP) receptors, plays an important role in the ecology of bacterial populations. This paper presents the first crystallographic analysis of the structure and ligand-induced conformational changes of the periplasmic tandem Per-Arnt-Sim (PAS) sensing domain (PTPSD) of a characterized MCP chemoreceptor. Analysis of the complex of the Campylobacter jejuni Tlp3 PTPSD with isoleucine (a chemoattractant) revealed that the PTPSD is a dimer in the crystal. The two ligand-binding sites are located in the membrane-distal PAS domains on the faces opposite to the dimer interface. Mutagenesis experiments show that the five strongly conserved residues that stabilize the main-chain moiety of isoleucine are essential for binding, suggesting that the mechanism by which this family of chemoreceptors recognizes amino acids is highly conserved. Although the fold and mode of ligand binding of the PTPSD are different from the aspartic acid receptor Tar, the structural analysis suggests that the PTPSDs of amino-acid chemoreceptors are also likely to signal by a piston displacement mechanism. The PTPSD fluctuates between piston (C-terminal helix) `up' and piston `down' states. Binding of an attractant to the distal PAS domain locks it in the closed form, weakening its association with the proximal domain and resulting in the transition of the latter into an open form, concomitant with a downward (towards the membrane) 4 Å piston displacement of the C-terminal helix. In vivo, this movement would generate a transmembrane signal by driving a downward displacement of the transmembrane helix 2 towards the cytoplasm.
Organizational Affiliation:
Department of Microbiology, Monash University, Clayton, Victoria 3800, Australia.