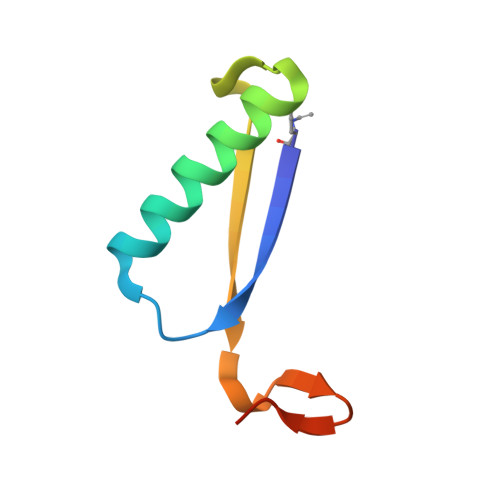
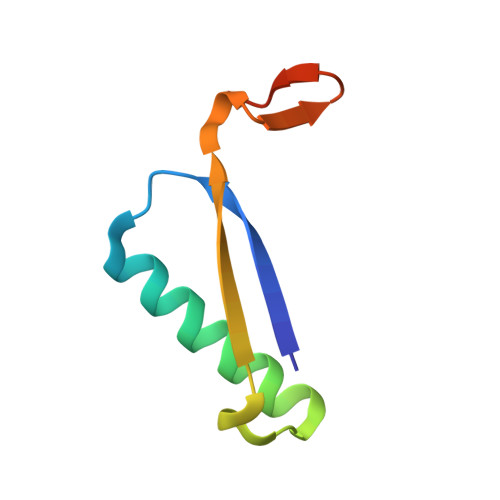
Evidence for the Formation of an Enamine Species during Aldol and Michael-type Addition Reactions Promiscuously Catalyzed by 4-Oxalocrotonate Tautomerase.
Poddar, H., Rahimi, M., Geertsema, E.M., Thunnissen, A.M., Poelarends, G.J.(2015) Chembiochem 16: 738-741
- PubMed: 25728471
- DOI: https://doi.org/10.1002/cbic.201402687
- Primary Citation of Related Structures:
4X19, 4X1C - PubMed Abstract:
The enzyme 4-oxalocrotonate tautomerase (4-OT), which has a catalytic N-terminal proline residue (Pro1), can promiscuously catalyze various carbon-carbon bond-forming reactions, including aldol condensation of acetaldehyde with benzaldehyde to yield cinnamaldehyde, and Michael-type addition of acetaldehyde to a wide variety of nitroalkenes to yield valuable γ-nitroaldehydes. To gain insight into how 4-OT catalyzes these unnatural reactions, we carried out exchange studies in D2 O, and X-ray crystallography studies. The former established that H-D exchange within acetaldehyde is catalyzed by 4-OT and that the Pro1 residue is crucial for this activity. The latter showed that Pro1 of 4-OT had reacted with acetaldehyde to give an enamine species. These results provide evidence of the mechanism of the 4-OT-catalyzed aldol and Michael-type addition reactions in which acetaldehyde is activated for nucleophilic addition by Pro1-dependent formation of an enamine intermediate.
Organizational Affiliation:
Department of Pharmaceutical Biology, Groningen Research Institute of Pharmacy, University of Groningen, Antonius Deusinglaan 1, 9713 AV Groningen (The Netherlands).