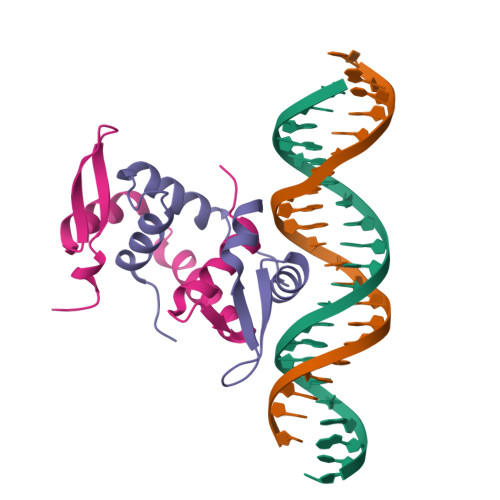
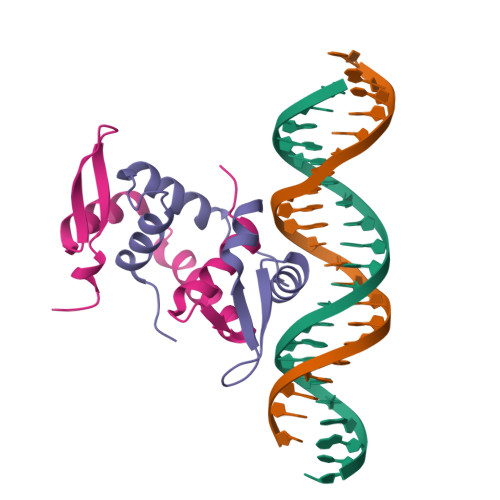
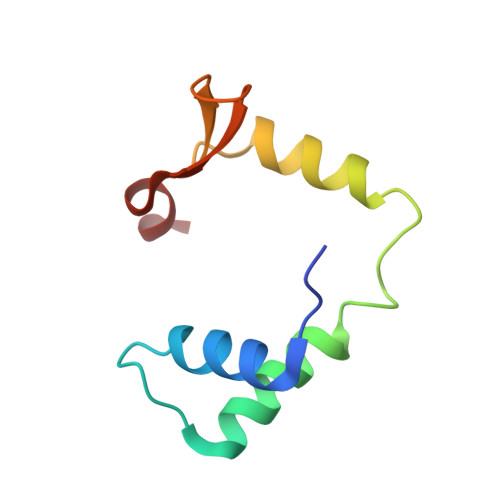
DNA binding by FOXP3 domain-swapped dimer suggests mechanisms of long-range chromosomal interactions.
Chen, Y., Chen, C., Zhang, Z., Liu, C.C., Johnson, M.E., Espinoza, C.A., Edsall, L.E., Ren, B., Zhou, X.J., Grant, S.F., Wells, A.D., Chen, L.(2015) Nucleic Acids Res 43: 1268-1282
- PubMed: 25567984
- DOI: https://doi.org/10.1093/nar/gku1373
- Primary Citation of Related Structures:
4WK8 - PubMed Abstract:
FOXP3 is a lineage-specific transcription factor that is required for regulatory T cell development and function. In this study, we determined the crystal structure of the FOXP3 forkhead domain bound to DNA. The structure reveals that FOXP3 can form a stable domain-swapped dimer to bridge DNA in the absence of cofactors, suggesting that FOXP3 may play a role in long-range gene interactions. To test this hypothesis, we used circular chromosome conformation capture coupled with high throughput sequencing (4C-seq) to analyze FOXP3-dependent genomic contacts around a known FOXP3-bound locus, Ptpn22. Our studies reveal that FOXP3 induces significant changes in the chromatin contacts between the Ptpn22 locus and other Foxp3-regulated genes, reflecting a mechanism by which FOXP3 reorganizes the genome architecture to coordinate the expression of its target genes. Our results suggest that FOXP3 mediates long-range chromatin interactions as part of its mechanisms to regulate specific gene expression in regulatory T cells.
Organizational Affiliation:
Laboratory of Structural Biology, Key Laboratory of Cancer Proteomics of Chinese Ministry of Health, XiangYa Hospital & State Key Laboratory of Medical Genetics, Central South University, Changsha, Hunan 410008, China Molecular and Computational Biology Program, Department of Biological Sciences, University of Southern California, Los Angeles, CA 90089, USA.