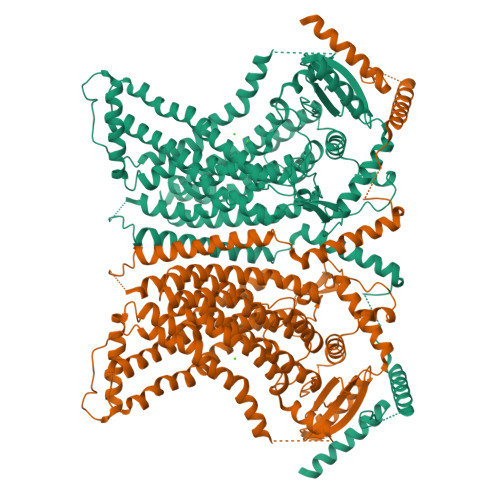
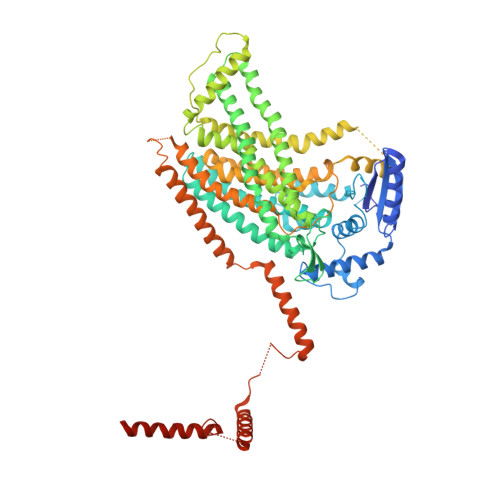
X-ray structure of a calcium-activated TMEM16 lipid scramblase.
Brunner, J.D., Lim, N.K., Schenck, S., Duerst, A., Dutzler, R.(2014) Nature 516: 207-212
- PubMed: 25383531
- DOI: https://doi.org/10.1038/nature13984
- Primary Citation of Related Structures:
4WIS, 4WIT - PubMed Abstract:
The TMEM16 family of proteins, also known as anoctamins, features a remarkable functional diversity. This family contains the long sought-after Ca(2+)-activated chloride channels as well as lipid scramblases and cation channels. Here we present the crystal structure of a TMEM16 family member from the fungus Nectria haematococca that operates as a Ca(2+)-activated lipid scramblase. Each subunit of the homodimeric protein contains ten transmembrane helices and a hydrophilic membrane-traversing cavity that is exposed to the lipid bilayer as a potential site of catalysis. This cavity harbours a conserved Ca(2+)-binding site located within the hydrophobic core of the membrane. Mutations of residues involved in Ca(2+) coordination affect both lipid scrambling in N. haematococca TMEM16 and ion conduction in the Cl(-) channel TMEM16A. The structure reveals the general architecture of the family and its mode of Ca(2+) activation. It also provides insight into potential scrambling mechanisms and serves as a framework to unravel the conduction of ions in certain TMEM16 proteins.
Organizational Affiliation:
Department of Biochemistry, University of Zurich, Winterthurerstrasse 190, CH-8057 Zurich, Switzerland.