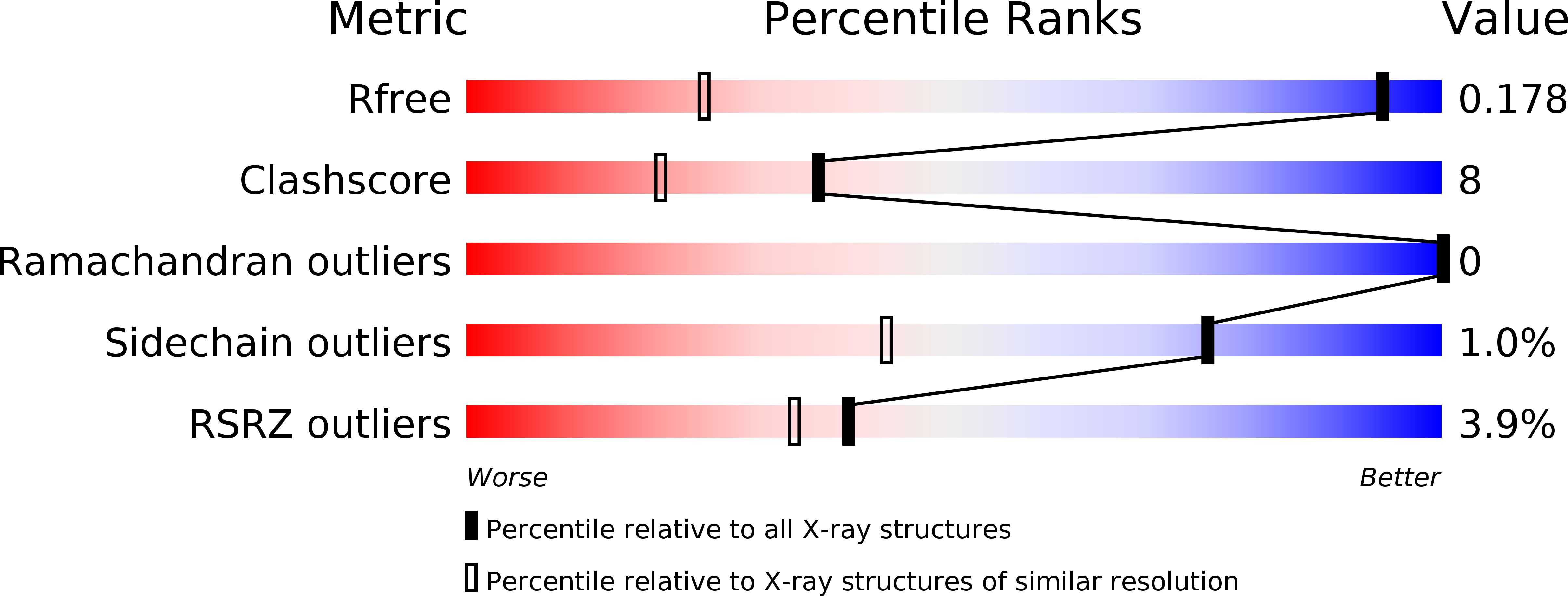
Rational Design and Asymmetric Synthesis of Potent and Neurotrophic Ligands for FK506-Binding Proteins (FKBPs).
Pomplun, S., Wang, Y., Kirschner, A., Kozany, C., Bracher, A., Hausch, F.(2015) Angew Chem Int Ed Engl 54: 345-348
- PubMed: 25412894
- DOI: https://doi.org/10.1002/anie.201408776
- Primary Citation of Related Structures:
4W9O, 4W9P, 4W9Q - PubMed Abstract:
To create highly efficient inhibitors for FK506-binding proteins, a new asymmetric synthesis for pro-(S)-C(5) -branched [4.3.1] aza-amide bicycles was developed. The key step of the synthesis is an HF-driven N-acyliminium cyclization. Functionalization of the C(5) moiety resulted in novel protein contacts with the psychiatric risk factor FKBP51, which led to a more than 280-fold enhancement in affinity. The most potent ligands facilitated the differentiation of N2a neuroblastoma cells with low nanomolar potency.
Organizational Affiliation:
Max Planck Institute of Psychiatry, Kraepelinstr. 2-10, 80804 Munich (Germany).