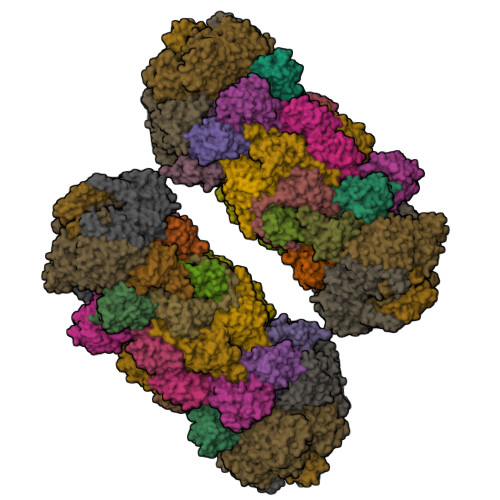
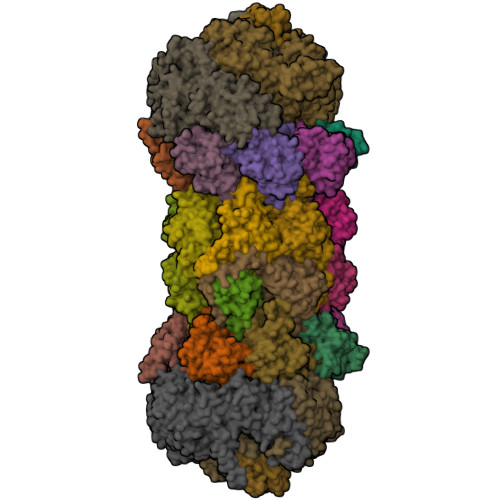
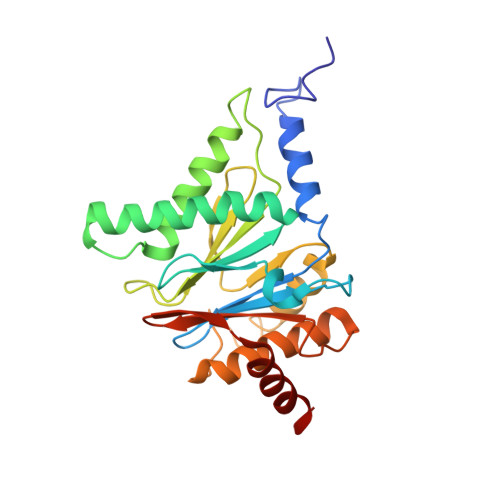
Structure of a Blm10 complex reveals common mechanisms for proteasome binding and gate opening.
Sadre-Bazzaz, K., Whitby, F.G., Robinson, H., Formosa, T., Hill, C.P.(2010) Mol Cell 37: 728-735
- PubMed: 20227375
- DOI: https://doi.org/10.1016/j.molcel.2010.02.002
- Primary Citation of Related Structures:
4V7O - PubMed Abstract:
The proteasome is an abundant protease that is critically important for numerous cellular pathways. Proteasomes are activated in vitro by three known classes of proteins/complexes, including Blm10/PA200. Here, we report a 3.4 A resolution crystal structure of a proteasome-Blm10 complex, which reveals that Blm10 surrounds the proteasome entry pore in the 1.2 MDa complex to form a largely closed dome that is expected to restrict access of potential substrates. This architecture and the observation that Blm10 induces a disordered proteasome gate structure challenge the assumption that Blm10 functions as an activator of proteolysis in vivo. The Blm10 C terminus binds in the same manner as seen for 11S activators and inferred for 19S/PAN activators and indicates a unified model for gate opening. We also demonstrate that Blm10 acts to maintain mitochondrial function. Consistent with the structural data, the C-terminal residues of Blm10 are needed for this activity.
Organizational Affiliation:
Department of Biochemistry, University of Utah School of Medicine, Salt Lake City, UT 84112-5650, USA.