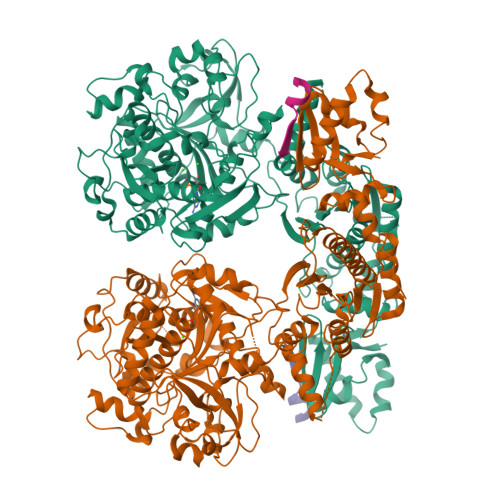
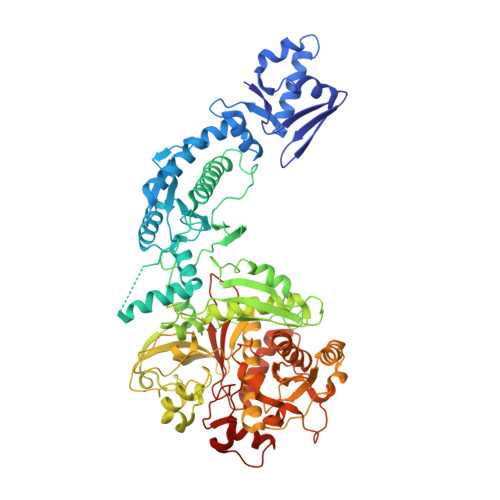
Structural Analysis of Leader Peptide Binding Enables Leader-Free Cyanobactin Processing.
Koehnke, J., Mann, G., Bent, A.F., Ludewig, H., Shirran, S., Botting, C., Lebl, T., Houssen, W.E., Jaspars, M., Naismith, J.H.(2015) Nat Chem Biol 11: 558
- PubMed: 26098679
- DOI: https://doi.org/10.1038/nchembio.1841
- Primary Citation of Related Structures:
4V1T, 4V1U, 4V1V - PubMed Abstract:
Regioselective modification of amino acids within the context of a peptide is common to a number of biosynthetic pathways, and many of the resulting products have potential as therapeutics. The ATP-dependent enzyme LynD heterocyclizes multiple cysteine residues to thiazolines within a peptide substrate. The enzyme requires the substrate to have a conserved N-terminal leader for full activity. Catalysis is almost insensitive to immediately flanking residues in the substrate, suggesting that recognition occurs distant from the active site. Nucleotide and peptide substrate co-complex structures of LynD reveal that the substrate leader peptide binds to and extends the β-sheet of a conserved domain of LynD, whereas catalysis is accomplished in another conserved domain. The spatial segregation of catalysis from recognition combines seemingly contradictory properties of regioselectivity and promiscuity, and it appears to be a conserved strategy in other peptide-modifying enzymes. A variant of LynD that efficiently processes substrates without a leader peptide has been engineered.
Organizational Affiliation:
BSRC, University of St Andrews, St Andrews, KY16 9RH.