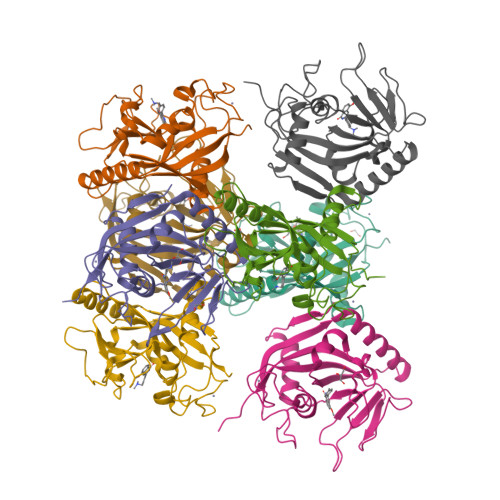
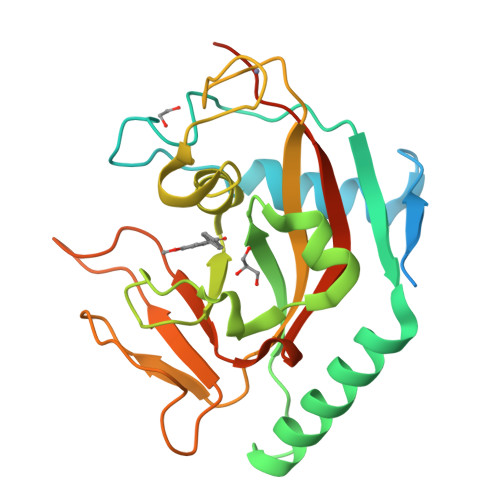
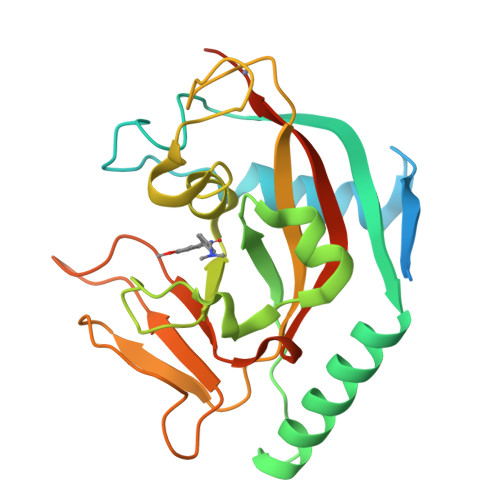
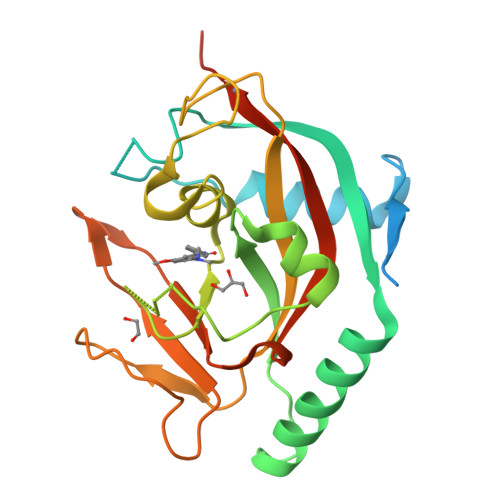
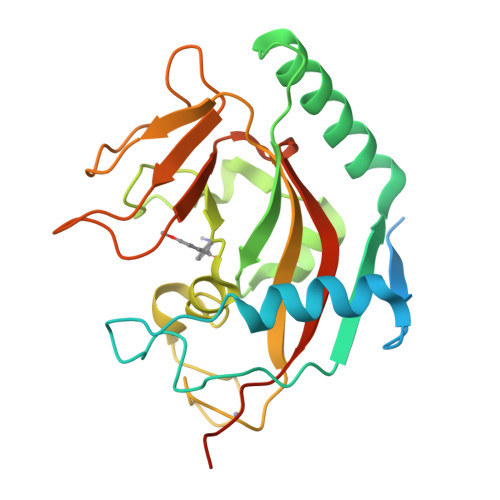
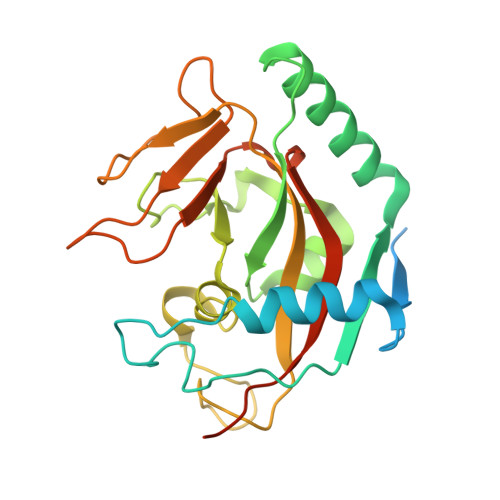
Design and Discovery of 3-Aryl-5-Substituted-Isoquinolin-1-Ones as Potent and Selective Tankyrase Inhibitors
Elliot, R.J., Jarvis, A., Rajasekaran, M.B., Menon, M., Bowers, L., Boffey, R., Bayford, M., Firth-Clark, S., Beevers, R., Aquil, R., Kirton, S.B., Niculescu-Duvaz, D., Fish, L., Lopes, F., Mcleary, R., Trindade, I., Vendrell, E., Munkonge, F., Porter, R., Perrior, T., Springer, C., Oliver, A.W., Pearl, L.H., Ashworth, A., Lord, C.J.(2015) Med Chem Res 6: 1687