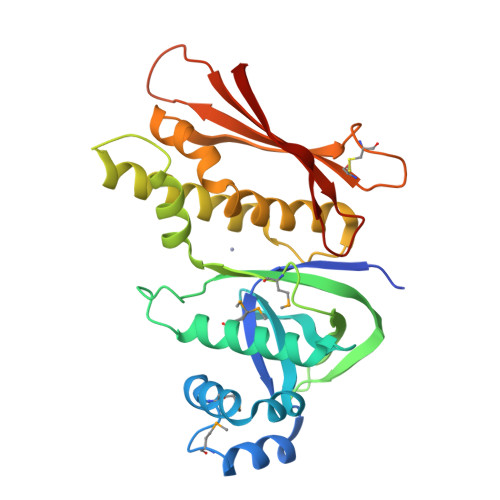
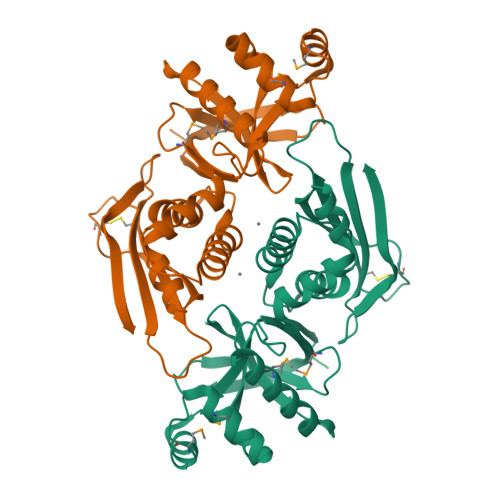
The Structure of the Cyanobactin Domain of Unknown Function from Patg in the Patellamide Gene Cluster
Mann, G., Koehnke, J., Bent, A.F., Graham, R., Schwarz-Linek, U., Naismith, J.H.(2014) Acta Crystallogr Sect F Struct Biol Cryst Commun 70: 1597
- PubMed: 25484206
- DOI: https://doi.org/10.1107/S2053230X1402425X
- Primary Citation of Related Structures:
4UVQ - PubMed Abstract:
Patellamides are members of the cyanobactin family of ribosomally synthesized and post-translationally modified cyclic peptide natural products, many of which, including some patellamides, are biologically active. A detailed mechanistic understanding of the biosynthetic pathway would enable the construction of a biotechnological `toolkit' to make novel analogues of patellamides that are not found in nature. All but two of the protein domains involved in patellamide biosynthesis have been characterized. The two domains of unknown function (DUFs) are homologous to each other and are found at the C-termini of the multi-domain proteins PatA and PatG. The domain sequence is found in all cyanobactin-biosynthetic pathways characterized to date, implying a functional role in cyanobactin biosynthesis. Here, the crystal structure of the PatG DUF domain is reported and its binding interactions with plausible substrates are investigated.
Organizational Affiliation:
Biomedical Sciences Research Complex, University of St Andrews, North Haugh, St Andrews, Fife KY16 8ST, Scotland.