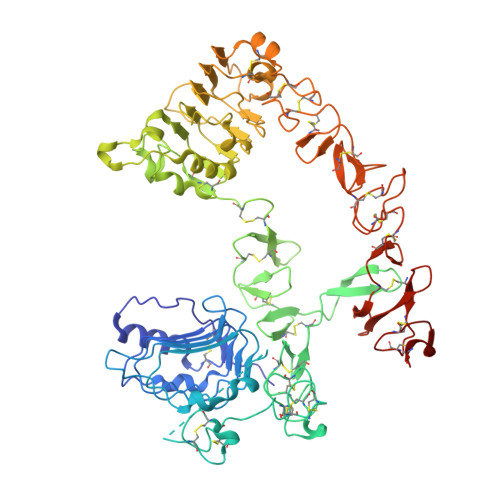
Gc1118, an Anti-Egfr Antibody with a Distinct Binding Epitope and Superior Inhibitory Activity Against High-Affinity Egfr Ligands.
Lim, Y., Yoo, J., Kim, M.S., Hur, M., Lee, E.H., Hur, H.S., Lee, J.C., Lee, S.N., Park, T.W., Lee, K., Chang, K.H., Kim, K., Kang, Y., Hong, K.W., Kim, S.H., Kim, Y.G., Yoon, Y., Nam, D.H., Yang, H., Kim, D.G., Cho, H.S., Won, J.(2016) Mol Cancer Ther 15: 251
- PubMed: 26586721
- DOI: https://doi.org/10.1158/1535-7163.MCT-15-0679
- Primary Citation of Related Structures:
4UV7 - PubMed Abstract:
The EGFR-targeted monoclonal antibodies are a valid therapeutic strategy for patients with metastatic colorectal cancer (mCRC). However, only a small subset of mCRC patients has therapeutic benefits and there are high demands for EGFR therapeutics with a broader patient pool and more potent efficacy. In this study, we report GC1118 exhibiting a different character in terms of binding epitope, affinity, mode of action, and efficacy from other anti-EGFR antibodies. Structural analysis of the EGFR-GC1118 crystal complex revealed that GC1118 recognizes linear, discrete N-terminal epitopes of domain III of EGFR, critical for EGF binding but not overlapping with those of other EGFR-targeted antibodies. GC1118 exhibited superior inhibitory activity against high-affinity EGFR ligands in terms of EGFR binding, triggering EGFR signaling, and proliferation compared with cetuximab and panitumumab. EGFR signaling driven by low-affinity ligands, on the contrary, was well inhibited by all the antibodies tested. GC1118 demonstrated robust antitumor activity in tumor xenografts with elevated expression of high-affinity ligands in vivo, whereas cetuximab did not. Considering the significant role of high-affinity EGFR ligands in modulating tumor microenvironment and inducing resistance to various cancer therapeutics, our study suggests a potential therapeutic advantage of GC1118 in terms of efficacy and a range of benefited patient pool. Mol Cancer Ther; 15(2); 251-63. ©2015 AACR.
Organizational Affiliation:
MOGAM Biotechnology Institute, Yongin, Gyeonggi-do, Republic of Korea.