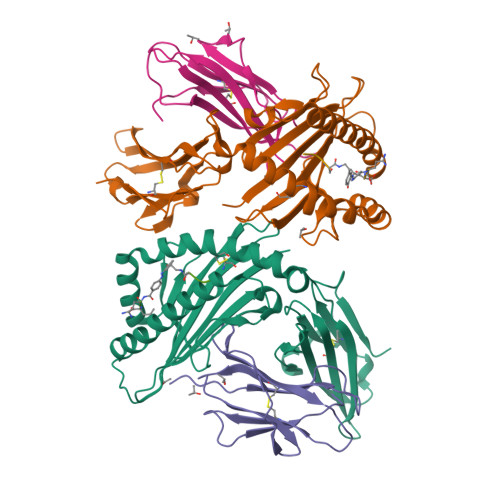
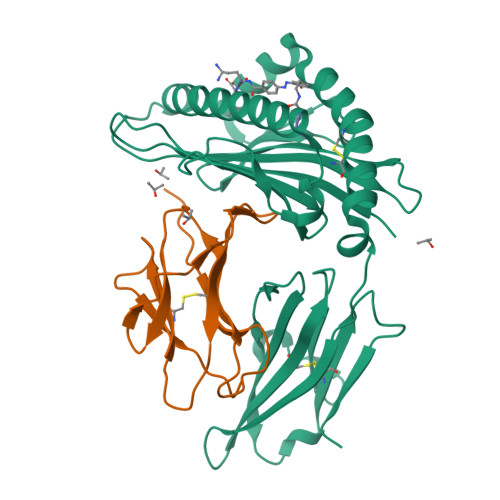
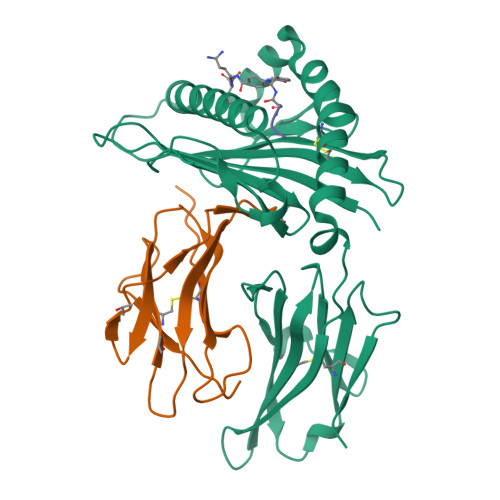
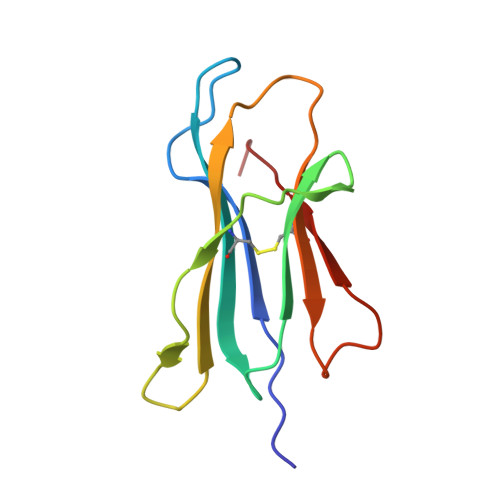
Bioorthogonal cleavage and exchange of major histocompatibility complex ligands by employing azobenzene-containing peptides.
Choo, J.A., Thong, S.Y., Yap, J., van Esch, W.J., Raida, M., Meijers, R., Lescar, J., Verhelst, S.H., Grotenbreg, G.M.(2014) Angew Chem Int Ed Engl 53: 13390-13394
- PubMed: 25348595
- DOI: https://doi.org/10.1002/anie.201406295
- Primary Citation of Related Structures:
4UQ2, 4UQ3 - PubMed Abstract:
Bioorthogonal cleavable linkers are attractive building blocks for compounds that can be manipulated to study biological and cellular processes. Sodium dithionite sensitive azobenzene-containing (Abc) peptides were applied for the temporary stabilization of recombinant MHC complexes, which can then be employed to generate libraries of MHC tetramers after exchange with a novel epitope. This technology represents an important tool for high-throughput studies of disease-specific T cell responses.
Organizational Affiliation:
Department of Microbiology and Immunology Programme, National University of Singapore (NUS), Life Sciences Institute, #03-05, 28 Medical Drive, 117456 (Singapore).