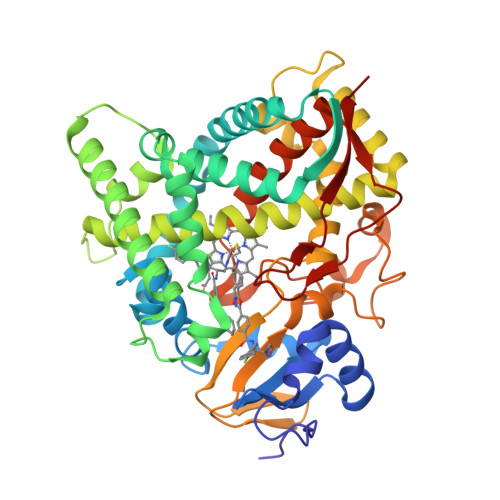
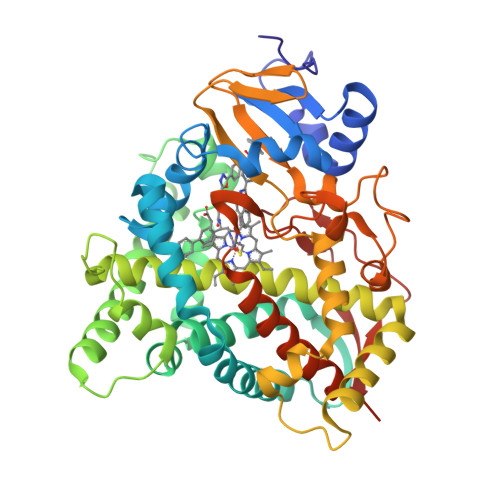
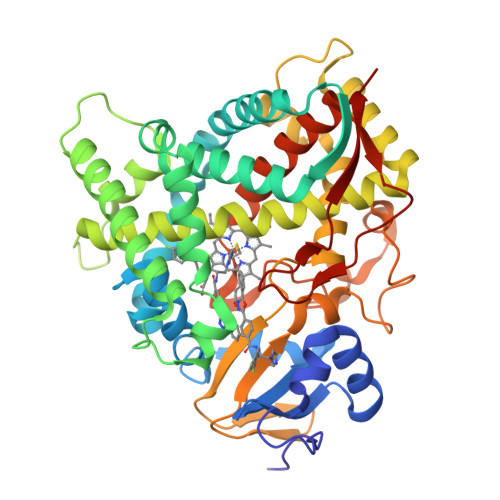
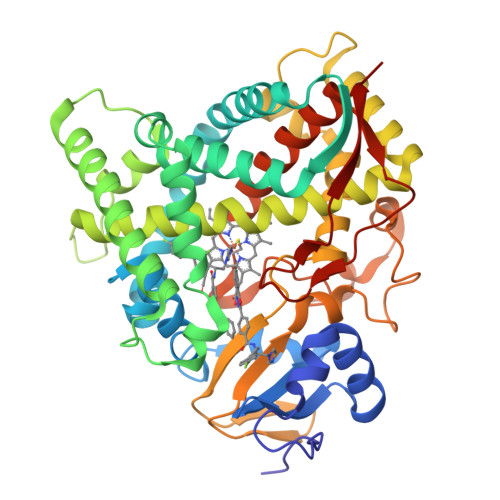
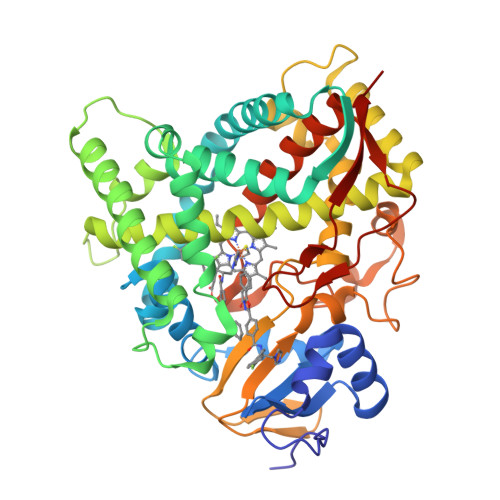
Human Sterol 14Alpha-Demethylase (Cyp51) as a Target for Anticancer Chemotherapy: Towards Structure-Aided Drug Design.
Hargrove, T.Y., Friggeri, L., Wawrzak, Z., Sivakumaran, S., Yazlovitskaya, E.M., Hiebert, S.W., Guengerich, F.P., Waterman, M.R., Lepesheva, G.I.(2016) J Lipid Res 57: 1552
- PubMed: 27313059
- DOI: https://doi.org/10.1194/jlr.M069229
- Primary Citation of Related Structures:
4UHI, 4UHL - PubMed Abstract:
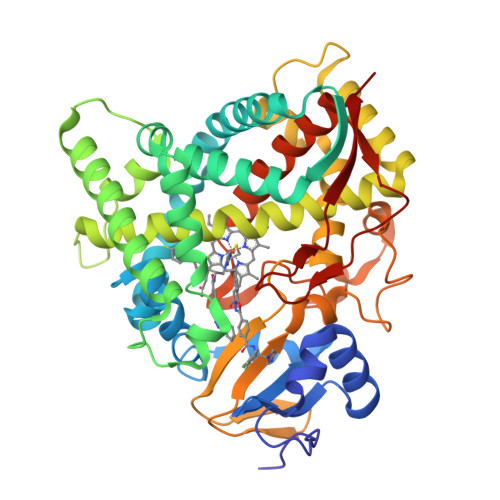
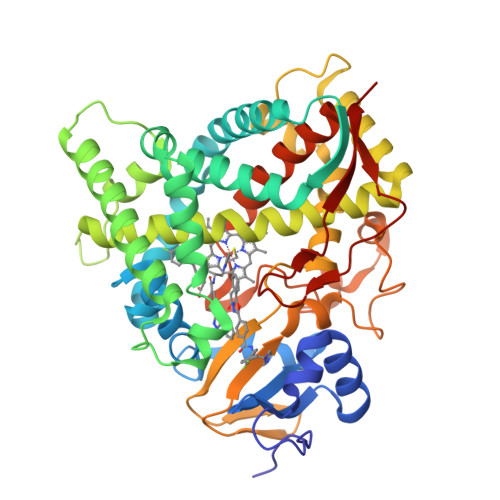
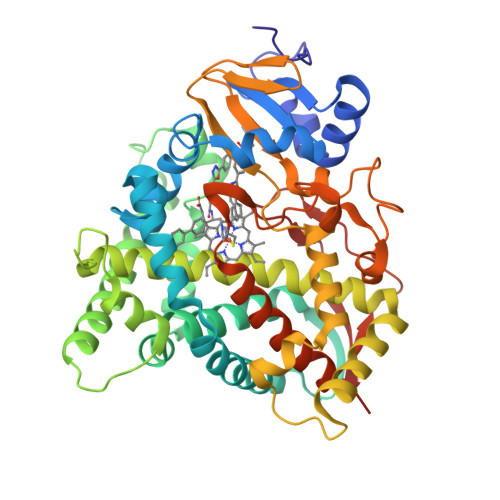
Rapidly multiplying cancer cells synthesize greater amounts of cholesterol to build their membranes. Cholesterol-lowering drugs (statins) are currently in clinical trials for anticancer chemotherapy. However, given at higher doses, statins cause serious side effects by inhibiting the formation of other biologically important molecules derived from mevalonate. Sterol 14α-demethylase (CYP51), which acts 10 steps downstream, is potentially a more specific drug target because this portion of the pathway is fully committed to cholesterol production. However, screening a variety of commercial and experimental inhibitors of microbial CYP51 orthologs revealed that most of them (including all clinical antifungals) weakly inhibit human CYP51 activity, even if they display high apparent spectral binding affinity. Only one relatively potent compound, (R)-N-(1-(3,4'-difluorobiphenyl-4-yl)-2-(1H-imidazol-1-yl)ethyl)-4-(5-phenyl-1,3,4-oxadiazol-2-yl)benzamide (VFV), was identified. VFV has been further tested in cellular experiments and found to decrease proliferation of different cancer cell types. The crystal structures of human CYP51-VFV complexes (2.0 and 2.5 Å) both display a 2:1 inhibitor/enzyme stoichiometry, provide molecular insights regarding a broader substrate profile, faster catalysis, and weaker susceptibility of human CYP51 to inhibition, and outline directions for the development of more potent inhibitors.
Organizational Affiliation:
Department of Biochemistry, Vanderbilt University School of Medicine, Nashville, TN 37232.