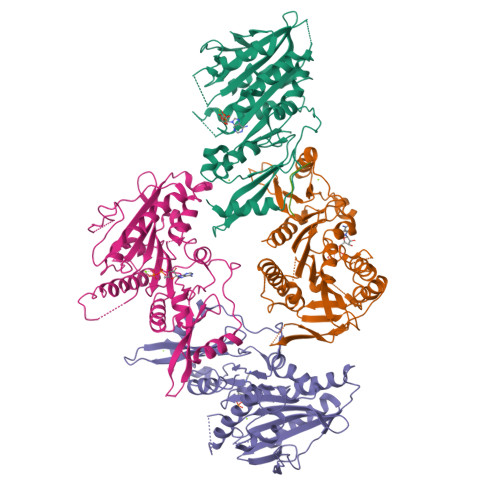
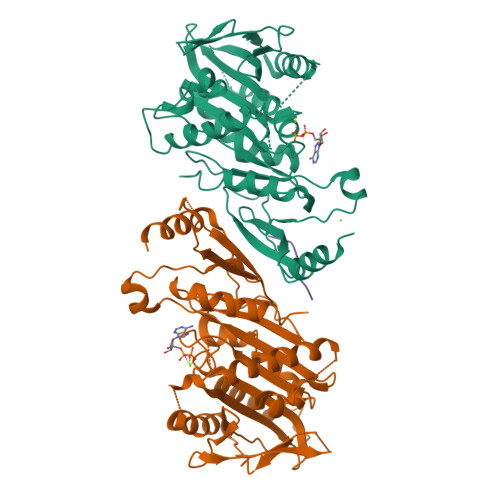
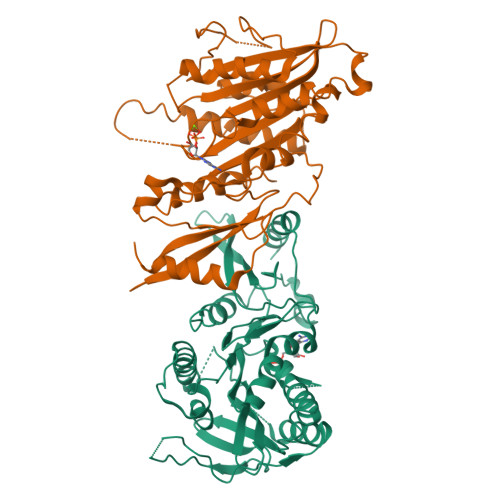
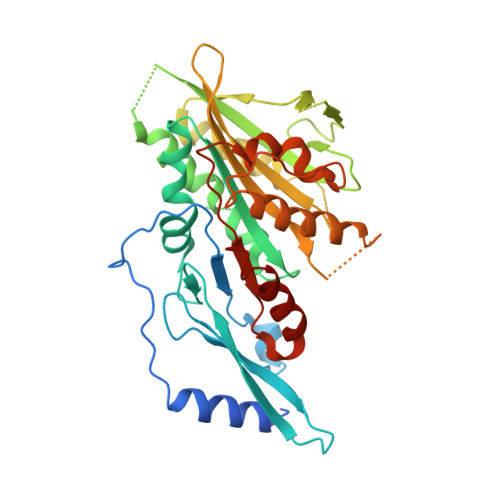
The C-terminal region of the motor protein MCAK controls its structure and activity through a conformational switch.
Talapatra, S.K., Harker, B., Welburn, J.P.(2015) Elife 4
- PubMed: 25915621
- DOI: https://doi.org/10.7554/eLife.06421
- Primary Citation of Related Structures:
4UBF - PubMed Abstract:
The precise regulation of microtubule dynamics is essential during cell division. The kinesin-13 motor protein MCAK is a potent microtubule depolymerase. The divergent non-motor regions flanking the ATPase domain are critical in regulating its targeting and activity. However, the molecular basis for the function of the non-motor regions within the context of full-length MCAK is unknown. Here, we determine the structure of MCAK motor domain bound to its regulatory C-terminus. Our analysis reveals that the MCAK C-terminus binds to two motor domains in solution and is displaced allosterically upon microtubule binding, which allows its robust accumulation at microtubule ends. These results demonstrate that MCAK undergoes long-range conformational changes involving its C-terminus during the soluble to microtubule-bound transition and that the C-terminus-motor interaction represents a structural intermediate in the MCAK catalytic cycle. Together, our work reveals intrinsic molecular mechanisms underlying the regulation of kinesin-13 activity.
Organizational Affiliation:
Wellcome Trust Centre for Cell Biology, School of Biological Sciences, University of Edinburgh, Edinburgh, United Kingdom.