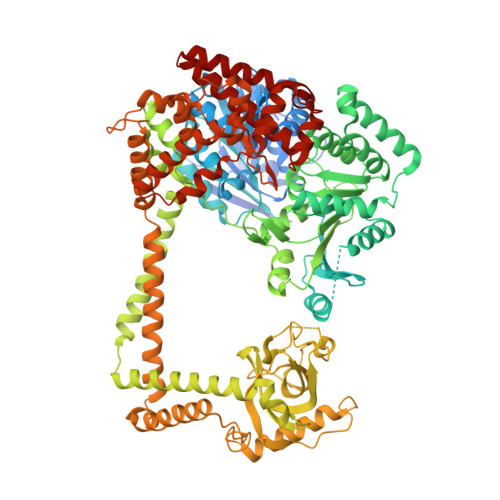
The Molecular Architecture of the TRAMP Complex Reveals the Organization and Interplay of Its Two Catalytic Activities.
Falk, S., Weir, J.R., Hentschel, J., Reichelt, P., Bonneau, F., Conti, E.(2014) Mol Cell 55: 856-867
- PubMed: 25175027
- DOI: https://doi.org/10.1016/j.molcel.2014.07.020
- Primary Citation of Related Structures:
4U4C - PubMed Abstract:
The TRAMP complex is involved in the nuclear surveillance and turnover of noncoding RNAs and intergenic transcripts. TRAMP is associated with the nuclear exosome and consists of a poly(A)polymerase subcomplex (Trf4-Air2) and a helicase (Mtr4). We found that N-terminal low-complexity regions of Trf4 and Air2 bind Mtr4 in a cooperative manner. The 2.4 Å resolution crystal structure of the corresponding ternary complex reveals how Trf4 and Air2 wrap around the DExH core of the helicase. Structure-based mutations on the DExH core impair binding to Trf4 and Air2, and also to Trf5 and Air1. The combination of structural, biochemical, and biophysical data suggests that the poly(A)polymerase core of Trf4-Air2 is positioned below the base of the helicase, where the unwound 3' end of an RNA substrate is expected to emerge. The results reveal conceptual similarities between the two major regulators of the exosome, the nuclear TRAMP and cytoplasmic Ski complexes.
Organizational Affiliation:
Structural Cell Biology Department, Max Planck Institute of Biochemistry, Am Klopferspitz 18, D-82152 Martinsried, Germany.