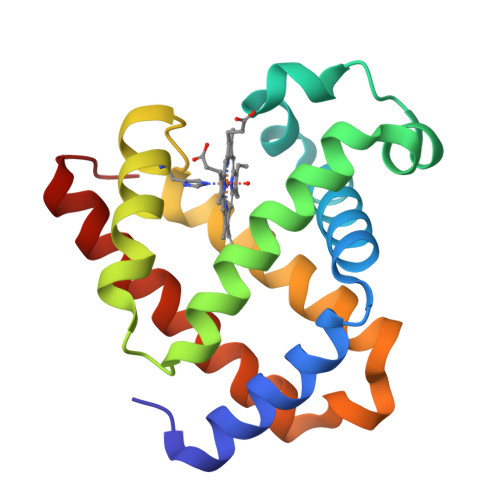
Systematic tuning of heme redox potentials and its effects on O2 reduction rates in a designed oxidase in myoglobin.
Bhagi-Damodaran, A., Petrik, I.D., Marshall, N.M., Robinson, H., Lu, Y.(2014) J Am Chem Soc 136: 11882-11885
- PubMed: 25076049
- DOI: https://doi.org/10.1021/ja5054863
- Primary Citation of Related Structures:
4TYX - PubMed Abstract:
Cytochrome c Oxidase (CcO) is known to catalyze the reduction of O2 to H2O efficiently with a much lower overpotential than most other O2 reduction catalysts. However, methods by which the enzyme fine-tunes the reduction potential (E°) of its active site and the corresponding influence on the O2 reduction activity are not well understood. In this work, we report systematic tuning of the heme E° in a functional model of CcO in myoglobin containing three histidines and one tyrosine in the distal pocket of heme. By removing hydrogen-bonding interactions between Ser92 and the proximal His ligand and a heme propionate, and increasing hydrophobicity of the heme pocket through Ser92Ala mutation, we have increased the heme E° from 95 ± 2 to 123 ± 3 mV. Additionally, replacing the native heme b in the CcO mimic with heme a analogs, diacetyl, monoformyl, and diformyl hemes, that posses electron-withdrawing groups, resulted in higher E° values of 175 ± 5, 210 ± 6, and 320 ± 10 mV, respectively. Furthermore, O2 consumption studies on these CcO mimics revealed a strong enhancement in O2 reduction rates with increasing heme E°. Such methods of tuning the heme E° through a combination of secondary sphere mutations and heme substitutions can be applied to tune E° of other heme proteins, allowing for comprehensive investigations of the relationship between E° and enzymatic activity.
Organizational Affiliation:
Department of Chemistry, University of Illinois, Urbana-Champaign , Urbana, Illinois 61801, United States.