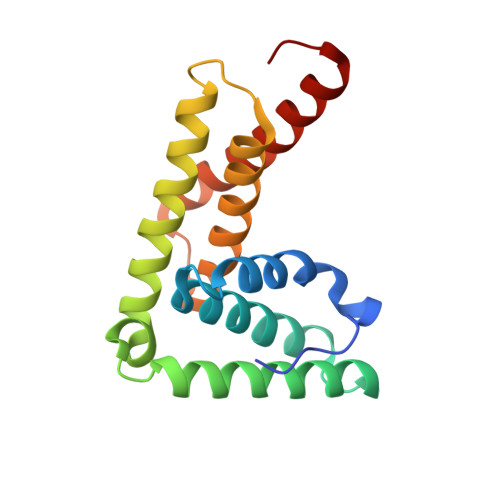
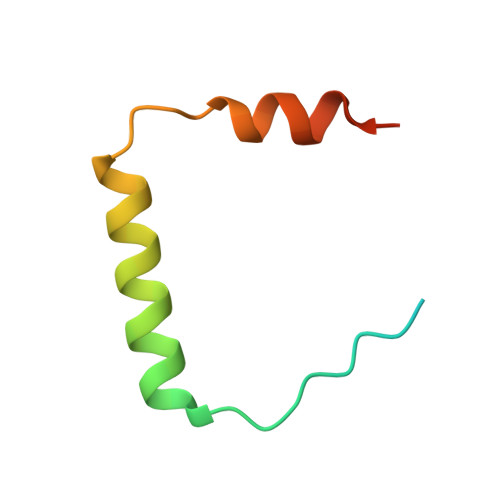
A Novel Mechanism of Regulating the ATPase VPS4 by Its Cofactor LIP5 and the Endosomal Sorting Complex Required for Transport (ESCRT)-III Protein CHMP5.
Vild, C.J., Li, Y., Guo, E.Z., Liu, Y., Xu, Z.(2015) J Biol Chem 290: 7291-7303
- PubMed: 25637630
- DOI: https://doi.org/10.1074/jbc.M114.616730
- Primary Citation of Related Structures:
4TXP, 4TXQ, 4TXR - PubMed Abstract:
Disassembly of the endosomal sorting complex required for transport (ESCRT) machinery from biological membranes is a critical final step in cellular processes that require the ESCRT function. This reaction is catalyzed by VPS4, an AAA-ATPase whose activity is tightly regulated by a host of proteins, including LIP5 and the ESCRT-III proteins. Here, we present structural and functional analyses of molecular interactions between human VPS4, LIP5, and the ESCRT-III proteins. The N-terminal domain of LIP5 (LIP5NTD) is required for LIP5-mediated stimulation of VPS4, and the ESCRT-III protein CHMP5 strongly inhibits the stimulation. Both of these observations are distinct from what was previously described for homologous yeast proteins. The crystal structure of LIP5NTD in complex with the MIT (microtubule-interacting and transport)-interacting motifs of CHMP5 and a second ESCRT-III protein, CHMP1B, was determined at 1 Å resolution. It reveals an ESCRT-III binding induced moderate conformational change in LIP5NTD, which results from insertion of a conserved CHMP5 tyrosine residue (Tyr(182)) at the core of LIP5NTD structure. Mutation of Tyr(182) partially relieves the inhibition displayed by CHMP5. Together, these results suggest a novel mechanism of VPS4 regulation in metazoans, where CHMP5 functions as a negative allosteric switch to control LIP5-mediated stimulation of VPS4.
Organizational Affiliation:
From the Life Sciences Institute and Department of Biological Chemistry, Medical School, University of Michigan, Ann Arbor, Michigan 48109.