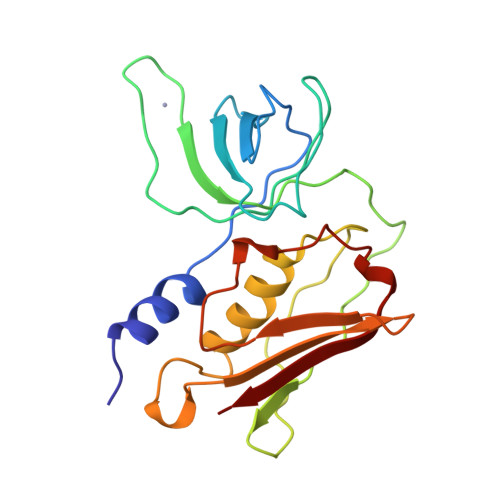
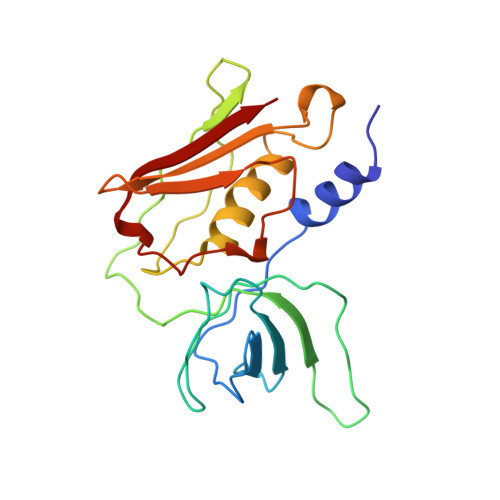
Refined structures of three crystal forms of toxic shock syndrome toxin-1 and of a tetramutant with reduced activity.
Prasad, G.S., Radhakrishnan, R., Mitchell, D.T., Earhart, C.A., Dinges, M.M., Cook, W.J., Schlievert, P.M., Ohlendorf, D.H.(1997) Protein Sci 6: 1220-1227
- PubMed: 9194182
- DOI: https://doi.org/10.1002/pro.5560060610
- Primary Citation of Related Structures:
2TSS, 3TSS, 4TSS, 5TSS - PubMed Abstract:
The structure of toxic shock syndrome toxin-1 (TSST-1), the causative agent in toxic shock syndrome, has been determined in three crystal forms. The three structural models have been refined to R-factors of 0.154, 0.150, and 0.198 at resolutions of 2.05 A, 2.90 A, and 2.75 A, respectively. One crystal form of TSST-1 contains a zinc ion bound between two symmetry-related molecules. Although not required for biological activity, zinc dramatically potentiates the mitogenicity of TSST-1 at very low concentrations. In addition, the structure of the tetramutant TSST-1H [T69I, Y80W, E132K, I140T], which is nonmitogenic and does not amplify endotoxin shock, has been determined and refined in a fourth crystal form (R-factor = 0.173 to 1.9 A resolution).
Organizational Affiliation:
Department of Biochemistry, Medical School, University of Minnesota, Minneapolis 55455, USA.