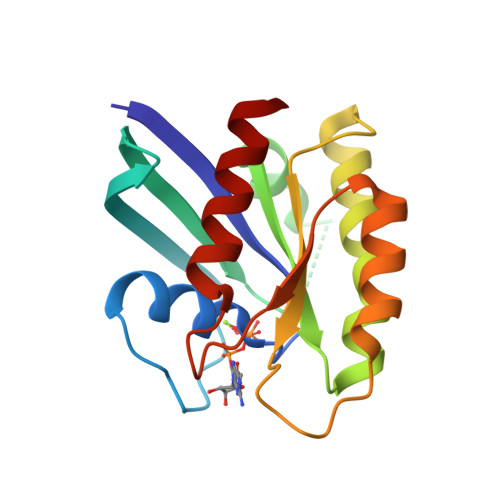
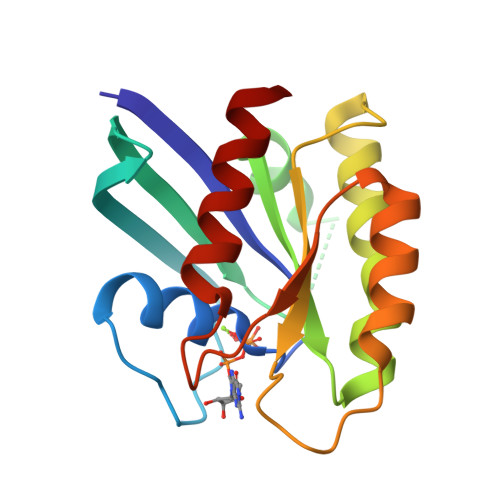
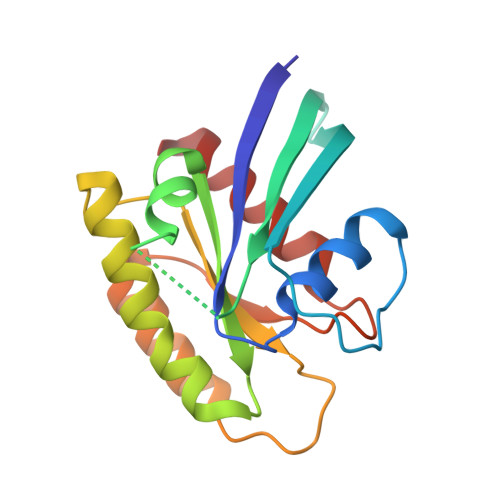
Neutron Crystal Structure of RAS GTPase Puts in Question the Protonation State of the GTP gamma-Phosphate.
Knihtila, R., Holzapfel, G., Weiss, K., Meilleur, F., Mattos, C.(2015) J Biological Chem 290: 31025-31036
- PubMed: 26515069
- DOI: https://doi.org/10.1074/jbc.M115.679860
- Primary Citation of Related Structures:
4RSG - PubMed Abstract:
RAS GTPase is a prototype for nucleotide-binding proteins that function by cycling between GTP and GDP, with hydrogen atoms playing an important role in the GTP hydrolysis mechanism. It is one of the most well studied proteins in the superfamily of small GTPases, which has representatives in a wide range of cellular functions. These proteins share a GTP-binding pocket with highly conserved motifs that promote hydrolysis to GDP. The neutron crystal structure of RAS presented here strongly supports a protonated γ-phosphate at physiological pH. This counters the notion that the phosphate groups of GTP are fully deprotonated at the start of the hydrolysis reaction, which has colored the interpretation of experimental and computational data in studies of the hydrolysis mechanism. The neutron crystal structure presented here puts in question our understanding of the pre-catalytic state associated with the hydrolysis reaction central to the function of RAS and other GTPases.
Organizational Affiliation:
From the Department of Chemistry and Chemical Biology, Northeastern University, Boston, Massachusetts 02115.