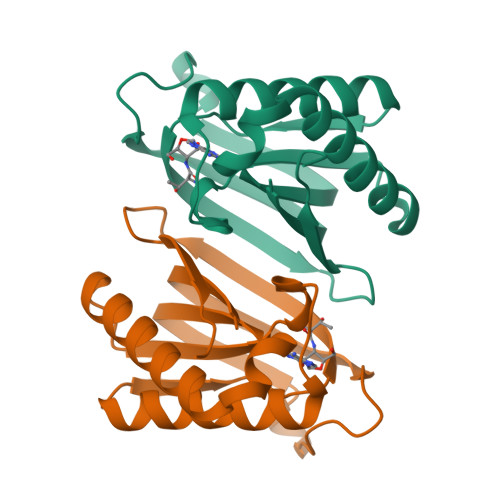
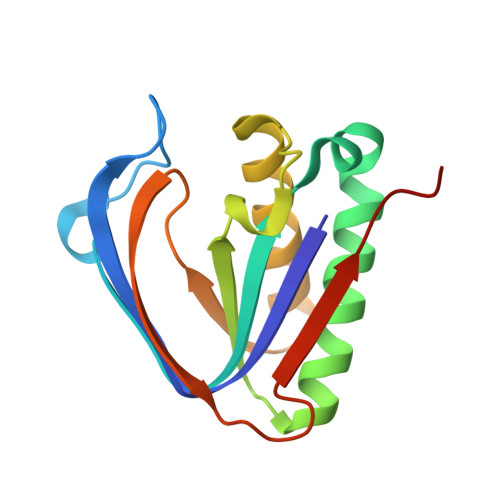
Specificity and catalysis hardwired at the RNA-protein interface in a translational proofreading enzyme.
Ahmad, S., Muthukumar, S., Kuncha, S.K., Routh, S.B., Yerabham, A.S., Hussain, T., Kamarthapu, V., Kruparani, S.P., Sankaranarayanan, R.(2015) Nat Commun 6: 7552-7552
- PubMed: 26113036
- DOI: https://doi.org/10.1038/ncomms8552
- Primary Citation of Related Structures:
4RR6, 4RR7, 4RR8, 4RR9, 4RRA, 4RRB, 4RRC, 4RRD, 4RRF, 4RRG, 4RRH, 4RRI, 4RRJ, 4RRK, 4RRL, 4RRM, 4RRQ, 4RRR - PubMed Abstract:
Proofreading modules of aminoacyl-tRNA synthetases are responsible for enforcing a high fidelity during translation of the genetic code. They use strategically positioned side chains for specifically targeting incorrect aminoacyl-tRNAs. Here, we show that a unique proofreading module possessing a D-aminoacyl-tRNA deacylase fold does not use side chains for imparting specificity or for catalysis, the two hallmark activities of enzymes. We show, using three distinct archaea, that a side-chain-stripped recognition site is fully capable of solving a subtle discrimination problem. While biochemical probing establishes that RNA plays the catalytic role, mechanistic insights from multiple high-resolution snapshots reveal that differential remodelling of the catalytic core at the RNA-peptide interface provides the determinants for correct proofreading activity. The functional crosstalk between RNA and protein elucidated here suggests how primordial enzyme functions could have emerged on RNA-peptide scaffolds before recruitment of specific side chains.
Organizational Affiliation:
Centre for Cellular and Molecular Biology, Council of Scientific and Industrial Research, Uppal Road, Hyderabad 500007, India.