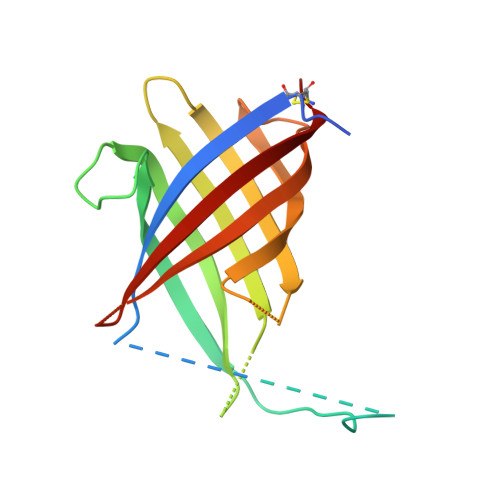
Trimeric Structure of (+)-Pinoresinol-forming Dirigent Protein at 1.95 angstrom Resolution with Three Isolated Active Sites.
Kim, K.W., Smith, C.A., Daily, M.D., Cort, J.R., Davin, L.B., Lewis, N.G.(2015) J Biol Chem 290: 1308-1318
- PubMed: 25411250
- DOI: https://doi.org/10.1074/jbc.M114.611780
- Primary Citation of Related Structures:
4REV - PubMed Abstract:
Control over phenoxy radical-radical coupling reactions in vivo in vascular plants was enigmatic until our discovery of dirigent proteins (DPs, from the Latin dirigere, to guide or align). The first three-dimensional structure of a DP ((+)-pinoresinol-forming DP, 1.95 Å resolution, rhombohedral space group H32)) is reported herein. It has a tightly packed trimeric structure with an eight-stranded β-barrel topology for each DP monomer. Each putative substrate binding and orientation coupling site is located on the trimer surface but too far apart for intermolecular coupling between sites. It is proposed that each site enables stereoselective coupling (using either two coniferyl alcohol radicals or a radical and a monolignol). Interestingly, there are six differentially conserved residues in DPs affording either the (+)- or (-)-antipodes in the vicinity of the putative binding site and region known to control stereoselectivity. DPs are involved in lignan biosynthesis, whereas dirigent domains/sites have been implicated in lignin deposition.
Organizational Affiliation:
From the Institute of Biological Chemistry, Washington State University, Pullman, Washington 99164-6340.