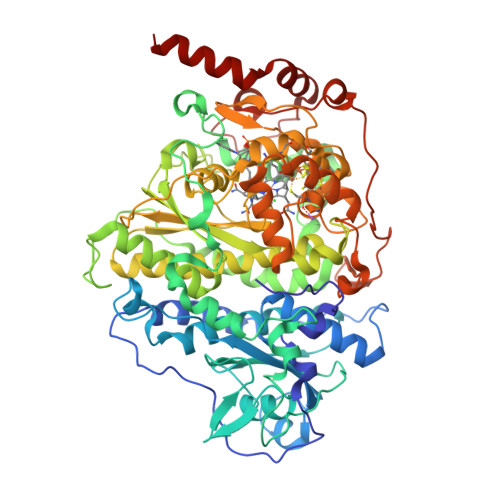
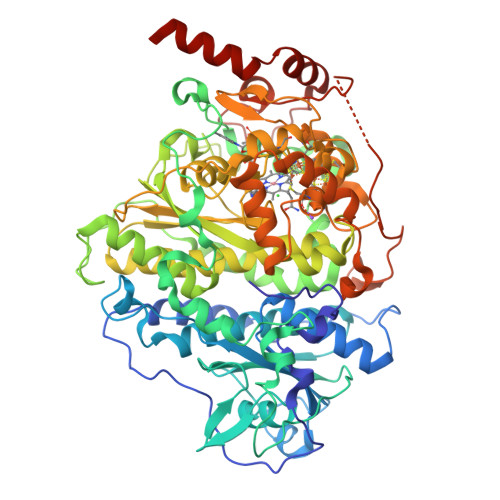
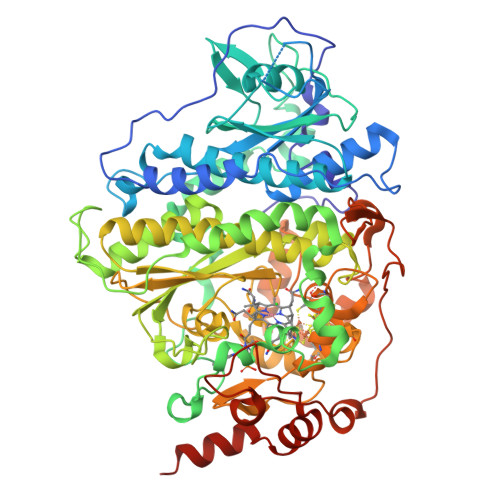
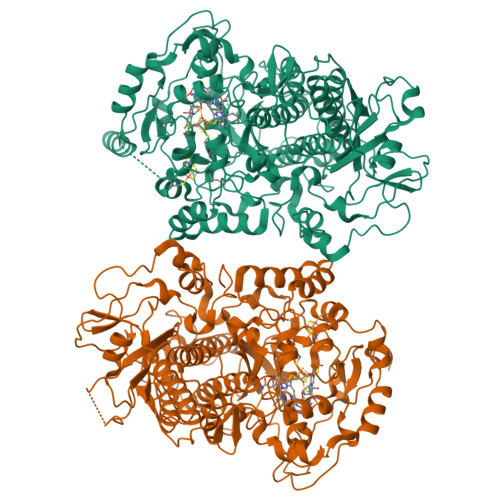
Reductive dehalogenase structure suggests a mechanism for B12-dependent dehalogenation.
Payne, K.A., Quezada, C.P., Fisher, K., Dunstan, M.S., Collins, F.A., Sjuts, H., Levy, C., Hay, S., Rigby, S.E., Leys, D.(2015) Nature 517: 513-516
- PubMed: 25327251
- DOI: https://doi.org/10.1038/nature13901
- Primary Citation of Related Structures:
4RAS - PubMed Abstract:
Organohalide chemistry underpins many industrial and agricultural processes, and a large proportion of environmental pollutants are organohalides. Nevertheless, organohalide chemistry is not exclusively of anthropogenic origin, with natural abiotic and biological processes contributing to the global halide cycle. Reductive dehalogenases are responsible for biological dehalogenation in organohalide respiring bacteria, with substrates including polychlorinated biphenyls or dioxins. Reductive dehalogenases form a distinct subfamily of cobalamin (B12)-dependent enzymes that are usually membrane associated and oxygen sensitive, hindering detailed studies. Here we report the characterization of a soluble, oxygen-tolerant reductive dehalogenase and, by combining structure determination with EPR (electron paramagnetic resonance) spectroscopy and simulation, show that a direct interaction between the cobalamin cobalt and the substrate halogen underpins catalysis. In contrast to the carbon-cobalt bond chemistry catalysed by the other cobalamin-dependent subfamilies, we propose that reductive dehalogenases achieve reduction of the organohalide substrate via halogen-cobalt bond formation. This presents a new model in both organohalide and cobalamin (bio)chemistry that will guide future exploitation of these enzymes in bioremediation or biocatalysis.
Organizational Affiliation:
Manchester Institute for Biotechnology, University of Manchester, Princess Street 131 M1 7DN Manchester, UK.