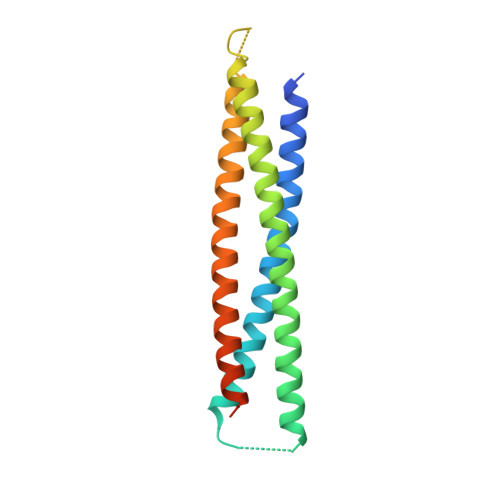
Single-chain protein mimetics of the N-terminal heptad-repeat region of gp41 with potential as anti-HIV-1 drugs.
Crespillo, S., Camara-Artigas, A., Casares, S., Morel, B., Cobos, E.S., Mateo, P.L., Mouz, N., Martin, C.E., Roger, M.G., El Habib, R., Su, B., Moog, C., Conejero-Lara, F.(2014) Proc Natl Acad Sci U S A 111: 18207-18212
- PubMed: 25489108
- DOI: https://doi.org/10.1073/pnas.1413592112
- Primary Citation of Related Structures:
4R61 - PubMed Abstract:
During HIV-1 fusion to the host cell membrane, the N-terminal heptad repeat (NHR) and the C-terminal heptad repeat (CHR) of the envelope subunit gp41 become transiently exposed and accessible to fusion inhibitors or Abs. In this process, the NHR region adopts a trimeric coiled-coil conformation that can be a target for therapeutic intervention. Here, we present an approach to rationally design single-chain protein constructs that mimic the NHR coiled-coil surface. The proteins were built by connecting with short loops two parallel NHR helices and an antiparallel one with the inverse sequence followed by engineering of stabilizing interactions. The constructs were expressed in Escherichia coli, purified with high yield, and folded as highly stable helical coiled coils. The crystal structure of one of the constructs confirmed the predicted fold and its ability to accurately mimic an exposed gp41 NHR surface. These single-chain proteins bound to synthetic CHR peptides with very high affinity, and furthermore, they showed broad inhibitory activity of HIV-1 fusion on various pseudoviruses and primary isolates.
- Departamento de Química Física e Instituto de Biotecnología, Facultad de Ciencias, Universidad de Granada, 18071 Granada, Spain;
Organizational Affiliation: