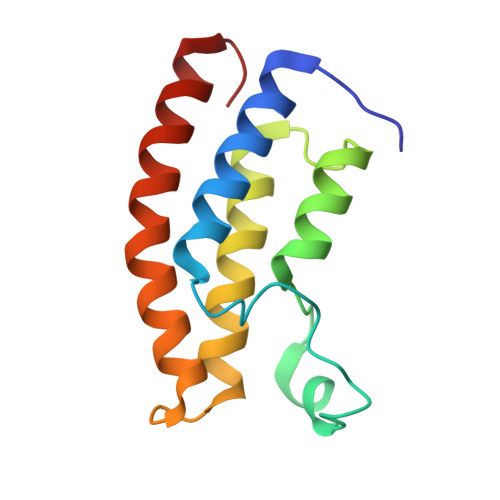
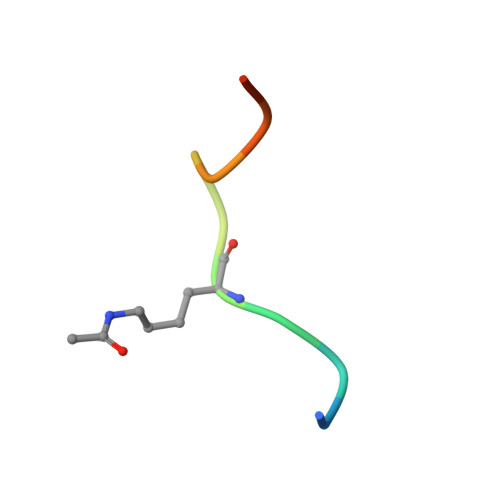
Structural insights into recognition of acetylated histone ligands by the BRPF1 bromodomain.
Lubula, M.Y., Eckenroth, B.E., Carlson, S., Poplawski, A., Chruszcz, M., Glass, K.C.(2014) FEBS Lett 588: 3844-3854
- PubMed: 25281266
- DOI: https://doi.org/10.1016/j.febslet.2014.09.028
- Primary Citation of Related Structures:
4QYD, 4QYL - PubMed Abstract:
Bromodomain-PHD finger protein 1 (BRPF1) is part of the MOZ HAT complex and contains a unique combination of domains typically found in chromatin-associated factors, which include plant homeodomain (PHD) fingers, a bromodomain and a proline-tryptophan-tryptophan-proline (PWWP) domain. Bromodomains are conserved structural motifs generally known to recognize acetylated histones, and the BRPF1 bromodomain preferentially selects for H2AK5ac, H4K12ac and H3K14ac. We solved the X-ray crystal structures of the BRPF1 bromodomain in complex with the H2AK5ac and H4K12ac histone peptides. Site-directed mutagenesis on residues in the BRPF1 bromodomain-binding pocket was carried out to investigate the contribution of specific amino acids on ligand binding. Our results provide critical insights into the molecular mechanism of ligand binding by the BRPF1 bromodomain, and reveal that ordered water molecules are an essential component driving ligand recognition.
Organizational Affiliation:
Department of Pharmaceutical Science, Albany College of Pharmacy and Health Sciences, Colchester, VT 05446, USA.