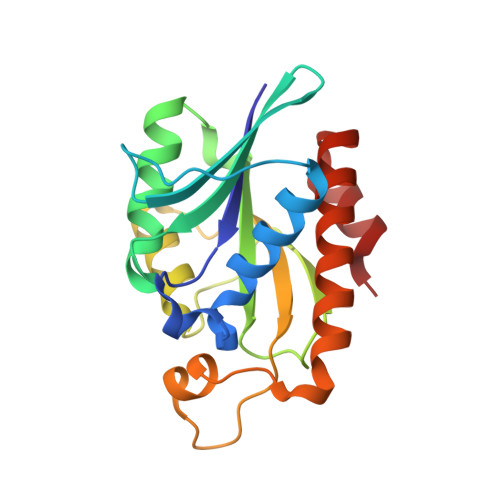
Crystal structure of peptidyl-tRNA hydrolase from a Gram-positive bacterium, Streptococcus pyogenes at 2.19 angstrom resolution shows the closed structure of the substrate-binding cleft.
Singh, A., Gautam, L., Sinha, M., Bhushan, A., Kaur, P., Sharma, S., Singh, T.P.(2014) FEBS Open Bio 4: 915-922
- PubMed: 25389518
- DOI: https://doi.org/10.1016/j.fob.2014.10.010
- Primary Citation of Related Structures:
4QT4 - PubMed Abstract:
Peptidyl-tRNA hydrolase (Pth) catalyses the release of tRNA and peptide components from peptidyl-tRNA molecules. Pth from a Gram-positive bacterium Streptococcus pyogenes (SpPth) was cloned, expressed, purified and crystallised. Three-dimensional structure of SpPth was determined by X-ray crystallography at 2.19 Å resolution. Structure determination showed that the asymmetric unit of the unit cell contained two crystallographically independent molecules, designated A and B. The superimposition of C(α) traces of molecules A and B showed an r.m.s. shift of 0.4 Å, indicating that the structures of two crystallographically independent molecules were identical. The polypeptide chain of SpPth adopted an overall α/β conformation. The substrate-binding cleft in SpPth is formed with three loops: the gate loop, Ile91-Leu102; the base loop, Gly108-Gly115; and the lid loop, Gly136-Gly150. Unlike in the structures of Pth from Gram-negative bacteria, the entry to the cleft in the structure of SpPth appeared to be virtually closed. However, the conformations of the active site residues were found to be similar.
Organizational Affiliation:
Department of Biophysics, All India Institute of Medical Sciences, New Delhi, India.