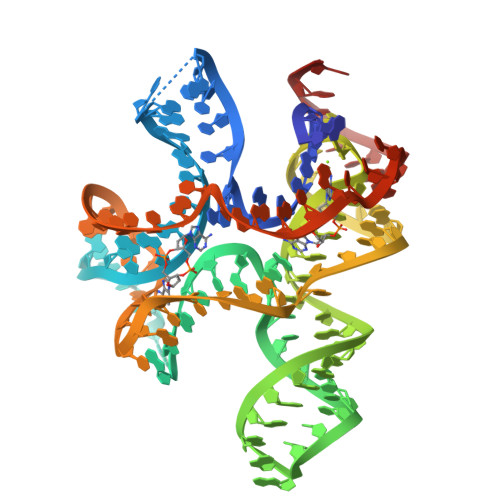
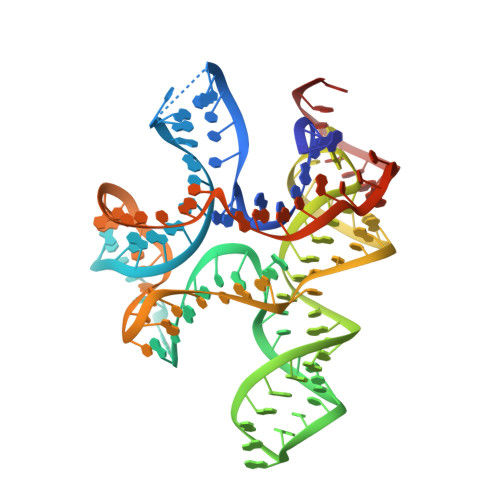
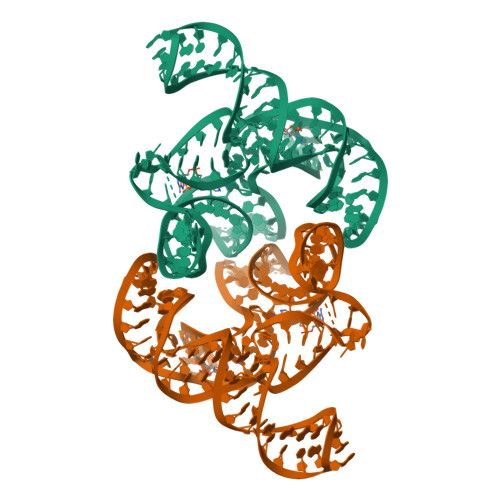c-di-AMP binds the ydaO riboswitch in two pseudo-symmetry-related pockets.
Ren, A., Patel, D.J.(2014) Nat Chem Biol 10: 780-786
- PubMed: 25086509
- DOI: https://doi.org/10.1038/nchembio.1606
- Primary Citation of Related Structures:
4QLM, 4QLN - PubMed Abstract:
The ydaO riboswitch, involved in sporulation, osmotic stress responses and cell wall metabolism, targets the second messenger cyclic-di-AMP with subnanomolar affinity. We have solved the structure of c-di-AMP bound to the Thermoanaerobacter tengcongensis ydaO riboswitch, thereby identifying a five-helical scaffold containing a zippered-up bubble, a pseudoknot and long-range tertiary base pairs. Highlights include the identification of two c-di-AMP binding pockets on the same face of the riboswitch, related by pseudo-two-fold symmetry, with potential for cross-talk between sites mediated by adjacently positioned base-stacking alignments connecting pockets. The adenine rings of bound c-di-AMP molecules are wedged between bases and stabilized by stacking, base-sugar and sugar-sugar intermolecular hydrogen bonding interactions. The structural studies are complemented by isothermal titration calorimetry-based binding studies of mutants mediating key tertiary intermolecular contacts. The T. tengcongensis ydaO riboswitch, like its Bacillus subtilis counterpart, most likely functions through a transcription termination mechanism, with the c-di-AMP bound state representing an 'off' switch.
Organizational Affiliation:
Structural Biology Program, Memorial Sloan-Kettering Cancer Center, New York, New York, USA.