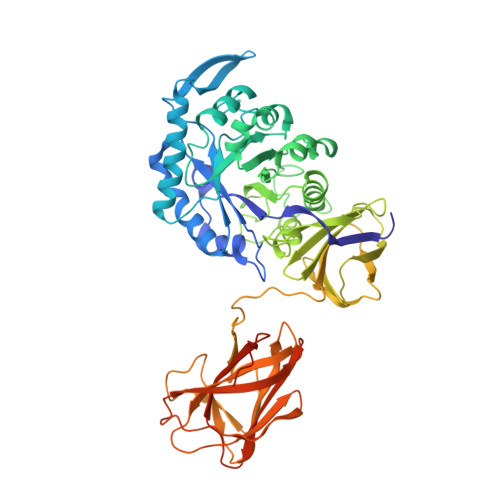
Structural Analysis of Glucuronoxylan-specific Xyn30D and Its Attached CBM35 Domain Gives Insights into the Role of Modularity in Specificity.
Sainz-Polo, M.A., Valenzuela, S.V., Gonzalez, B., Pastor, F.I., Sanz-Aparicio, J.(2014) J Biol Chem 289: 31088-31101
- PubMed: 25202007
- DOI: https://doi.org/10.1074/jbc.M114.597732
- Primary Citation of Related Structures:
4QAW, 4QB1, 4QB2, 4QB6 - PubMed Abstract:
Glucuronoxylanase Xyn30D is a modular enzyme containing a family 30 glycoside hydrolase catalytic domain and an attached carbohydrate binding module of the CBM35 family. We present here the three-dimensional structure of the full-length Xyn30D at 2.4 Å resolution. The catalytic domain folds into an (α/β)8 barrel with an associated β-structure, whereas the attached CBM35 displays a jellyroll β-sandwich including two calcium ions. Although both domains fold in an independent manner, the linker region makes polar interactions with the catalytic domain, allowing a moderate flexibility. The ancillary Xyn30D-CBM35 domain has been expressed and crystallized, and its binding abilities have been investigated by soaking experiments. Only glucuronic acid-containing ligands produced complexes, and their structures have been solved. A calcium-dependent glucuronic acid binding site shows distinctive structural features as compared with other uronic acid-specific CBM35s, because the presence of two aromatic residues delineates a wider pocket. The nonconserved Glu(129) makes a bidentate link to calcium and defines region E, previously identified as specificity hot spot. The molecular surface of Xyn30D-CBM35 shows a unique stretch of negative charge distribution extending from its binding pocket that might indicate some oriented interaction with its target substrate. The binding ability of Xyn30D-CBM35 to different xylans was analyzed by affinity gel electrophoresis. Some binding was observed with rye glucuronoarabinoxylan in presence of calcium chelating EDTA, which would indicate that Xyn30D-CBM35 might establish interaction to other components of xylan, such as arabinose decorations of glucuronoarabinoxylan. A role in depolymerization of highly substituted chemically complex xylans is proposed.
Organizational Affiliation:
From the Departamento de Cristalografía y Biología Estructural, Instituto de Química-Física Rocasolano, CSIC, 28006 Madrid, Spain and.