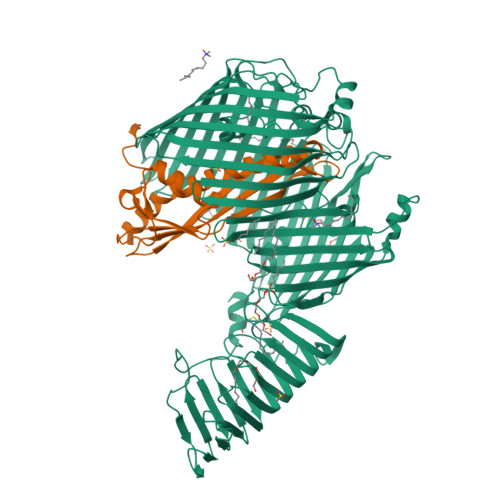
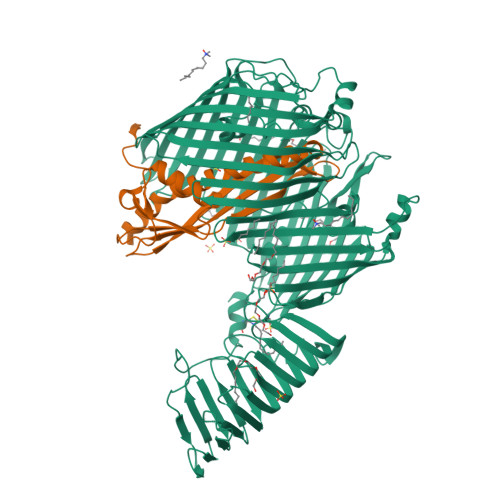
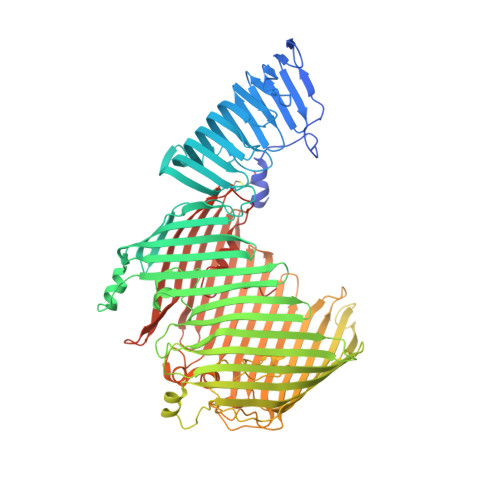
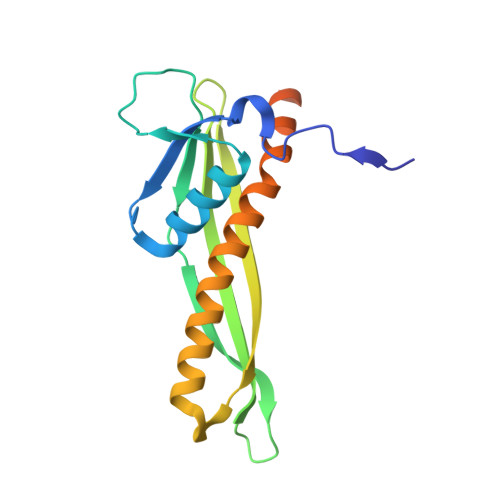
Structural basis for lipopolysaccharide insertion in the bacterial outer membrane.
Qiao, S., Luo, Q., Zhao, Y., Zhang, X.C., Huang, Y.(2014) Nature 511: 108-111
- PubMed: 24990751
- DOI: https://doi.org/10.1038/nature13484
- Primary Citation of Related Structures:
4Q35 - PubMed Abstract:
One of the fundamental properties of biological membranes is the asymmetric distribution of membrane lipids. In Gram-negative bacteria, the outer leaflet of the outer membrane is composed predominantly of lipopolysaccharides (LPS). The export of LPS requires seven essential lipopolysaccharide transport (Lpt) proteins to move LPS from the inner membrane, through the periplasm to the surface. Of the seven Lpt proteins, the LptD-LptE complex is responsible for inserting LPS into the external leaflet of the outer membrane. Here we report the crystal structure of the ∼110-kilodalton membrane protein complex LptD-LptE from Shigella flexneri at 2.4 Å resolution. The structure reveals an unprecedented two-protein plug-and-barrel architecture with LptE embedded into a 26-stranded β-barrel formed by LptD. Importantly, the secondary structures of the first two β-strands are distorted by two proline residues, weakening their interactions with neighbouring β-strands and creating a potential portal on the barrel wall that could allow lateral diffusion of LPS into the outer membrane. The crystal structure of the LptD-LptE complex opens the door to new antibiotic strategies targeting the bacterial outer membrane.
Organizational Affiliation:
1] National Laboratory of Biomacromolecules, National Center of Protein Science-Beijing, Institute of Biophysics, Chinese Academy of Sciences, Beijing 100101, China [2] University of Chinese Academy of Sciences, Beijing 100101, China.