Agrobacterium uses a unique ligand-binding mode for trapping opines and acquiring a competitive advantage in the niche construction on plant host.
Lang, J., Vigouroux, A., Planamente, S., El Sahili, A., Blin, P., Aumont-Nicaise, M., Dessaux, Y., Morera, S., Faure, D.(2014) PLoS Pathog 10: e1004444-e1004444
- PubMed: 25299655
- DOI: https://doi.org/10.1371/journal.ppat.1004444
- Primary Citation of Related Structures:
4P0I, 4POW, 4PP0, 5OVZ - PubMed Abstract:
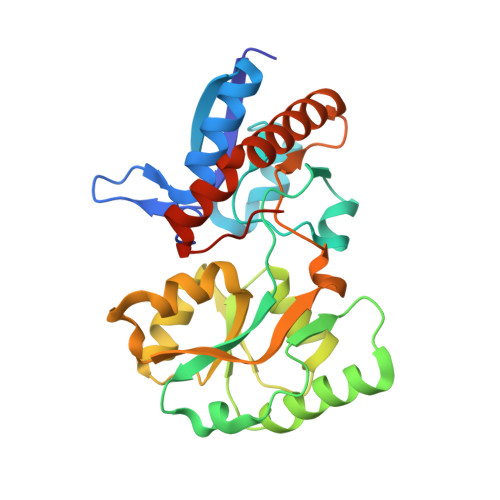
By modifying the nuclear genome of its host, the plant pathogen Agrobacterium tumefaciens induces the development of plant tumours in which it proliferates. The transformed plant tissues accumulate uncommon low molecular weight compounds called opines that are growth substrates for A. tumefaciens. In the pathogen-induced niche (the plant tumour), a selective advantage conferred by opine assimilation has been hypothesized, but not experimentally demonstrated. Here, using genetics and structural biology, we deciphered how the pathogen is able to bind opines and use them to efficiently compete in the plant tumour. We report high resolution X-ray structures of the periplasmic binding protein (PBP) NocT unliganded and liganded with the opine nopaline (a condensation product of arginine and α-ketoglurate) and its lactam derivative pyronopaline. NocT exhibited an affinity for pyronopaline (K(D) of 0.6 µM) greater than that for nopaline (KD of 3.7 µM). Although the binding-mode of the arginine part of nopaline/pyronopaline in NocT resembled that of arginine in other PBPs, affinity measurement by two different techniques showed that NocT did not bind arginine. In contrast, NocT presented specific residues such as M117 to stabilize the bound opines. NocT relatives that exhibit the nopaline/pyronopaline-binding mode were only found in genomes of the genus Agrobacterium. Transcriptomics and reverse genetics revealed that A. tumefaciens uses the same pathway for assimilating nopaline and pyronopaline. Fitness measurements showed that NocT is required for a competitive colonization of the plant tumour by A. tumefaciens. Moreover, even though the Ti-plasmid conjugal transfer was not regulated by nopaline, the competitive advantage gained by the nopaline-assimilating Ti-plasmid donors led to a preferential horizontal propagation of this Ti-plasmid amongst the agrobacteria colonizing the plant-tumour niche. This work provided structural and genetic evidences to support the niche construction paradigm in bacterial pathogens.
Organizational Affiliation:
Institut des Sciences du Végétal (ISV), UPR2355, CNRS, Saclay Plant sciences, Gif-sur-Yvette, France.