Structural and Thermodynamic Insights into Chitooligosaccharide Binding to Human Cartilage Chitinase 3-like Protein 2 (CHI3L2 or YKL-39).
Ranok, A., Wongsantichon, J., Robinson, R.C., Suginta, W.(2015) J Biological Chem 290: 2617-2629
- PubMed: 25477513
- DOI: https://doi.org/10.1074/jbc.M114.588905
- Primary Citation of Related Structures:
4P8U, 4P8V, 4P8W, 4P8X - PubMed Abstract:
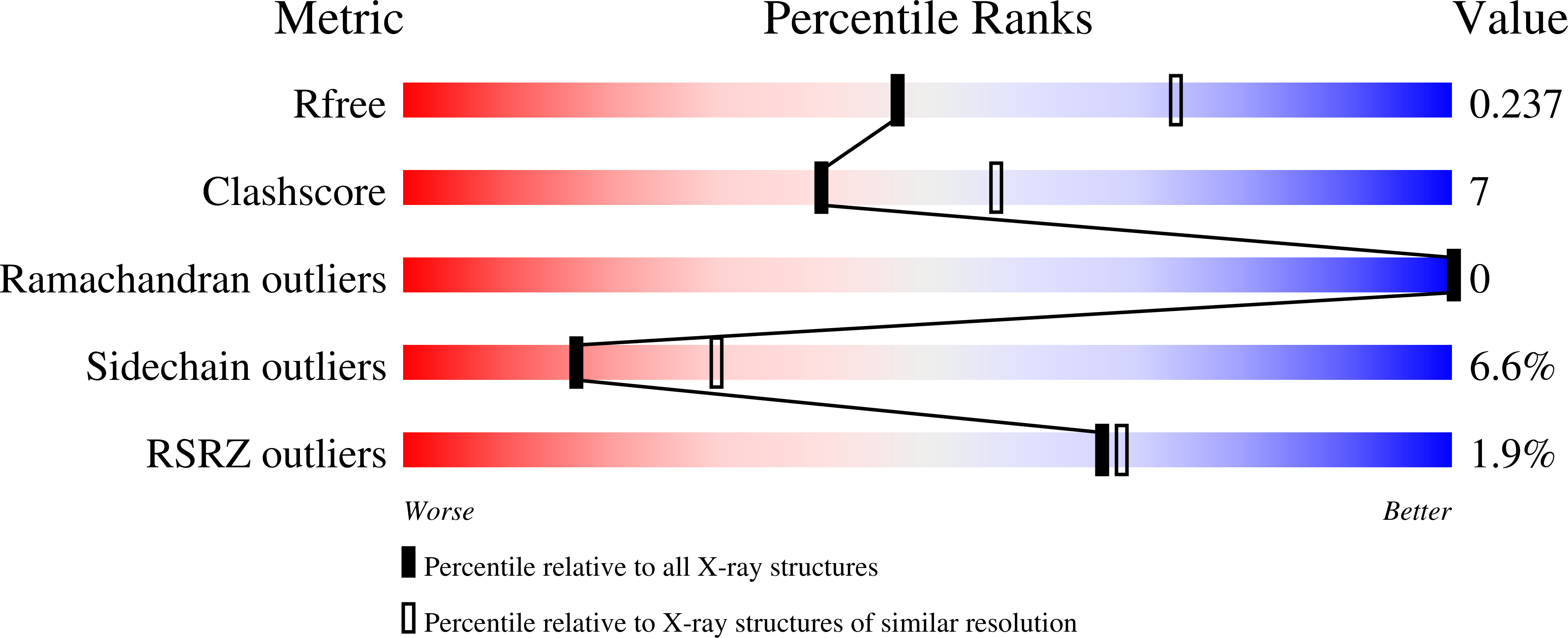
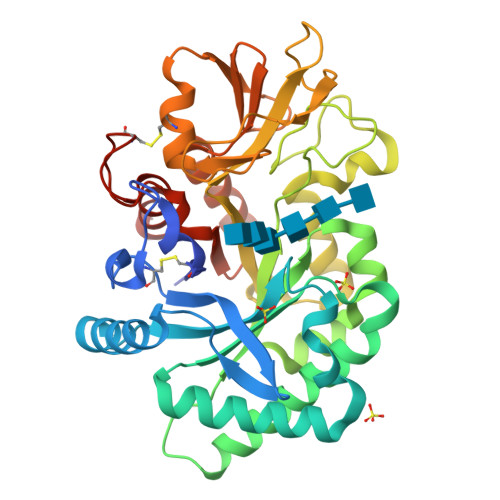
Four crystal structures of human YKL-39 were solved in the absence and presence of chitooligosaccharides. The structure of YKL-39 comprises a major (β/α)8 triose-phosphate isomerase barrel domain and a small α + β insertion domain. Structural analysis demonstrates that YKL-39 interacts with chitooligosaccharides through hydrogen bonds and hydrophobic interactions. The binding of chitin fragments induces local conformational changes that facilitate tight binding. Compared with other GH-18 members, YKL-39 has the least extended chitin-binding cleft, containing five subsites for sugars, namely (-3)(-2)(-1)(+1)(+2), with Trp-360 playing a prominent role in the sugar-protein interactions at the center of the chitin-binding cleft. Evaluation of binding affinities obtained from isothermal titration calorimetry and intrinsic fluorescence spectroscopy suggests that YKL-39 binds to chitooligosaccharides with Kd values in the micromolar concentration range and that the binding energies increase with the chain length. There were no significant differences between the Kd values of chitopentaose and chitohexaose, supporting the structural evidence for the five binding subsite topology. Thermodynamic analysis indicates that binding of chitooligosaccharide to YKL-39 is mainly driven by enthalpy.
Organizational Affiliation:
From the Biochemistry-Electrochemistry Research Unit, School of Biochemistry, Institute of Science, Suranaree University of Technology, Nakhon Ratchasima 30000, Thailand.