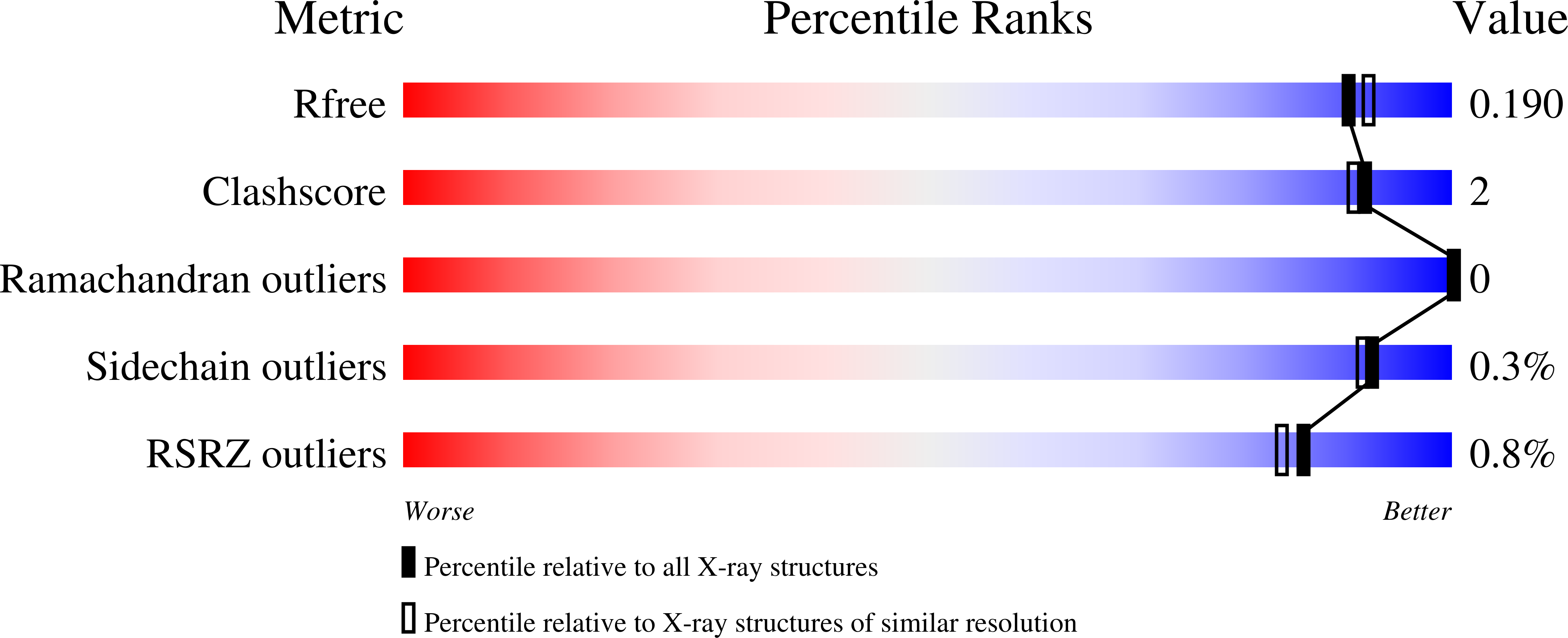
Modification and periplasmic translocation of the biofilm exopolysaccharide poly-beta-1,6-N-acetyl-D-glucosamine.
Little, D.J., Li, G., Ing, C., DiFrancesco, B.R., Bamford, N.C., Robinson, H., Nitz, M., Pomes, R., Howell, P.L.(2014) Proc Natl Acad Sci U S A 111: 11013-11018
- PubMed: 24994902
- DOI: https://doi.org/10.1073/pnas.1406388111
- Primary Citation of Related Structures:
4P7L, 4P7N, 4P7O, 4P7Q, 4P7R - PubMed Abstract:
Poly-β-1,6-N-acetyl-D-glucosamine (PNAG) is an exopolysaccharide produced by a wide variety of medically important bacteria. Polyglucosamine subunit B (PgaB) is responsible for the de-N-acetylation of PNAG, a process required for polymer export and biofilm formation. PgaB is located in the periplasm and likely bridges the inner membrane synthesis and outer membrane export machinery. Here, we present structural, functional, and molecular simulation data that suggest PgaB associates with PNAG continuously during periplasmic transport. We show that the association of PgaB's N- and C-terminal domains forms a cleft required for the binding and de-N-acetylation of PNAG. Molecular dynamics (MD) simulations of PgaB show a binding preference for N-acetylglucosamine (GlcNAc) to the N-terminal domain and glucosammonium to the C-terminal domain. Continuous ligand binding density is observed that extends around PgaB from the N-terminal domain active site to an electronegative groove on the C-terminal domain that would allow for a processive mechanism. PgaB's C-terminal domain (PgaB310-672) directly binds PNAG oligomers with dissociation constants of ∼1-3 mM, and the structures of PgaB310-672 in complex with β-1,6-(GlcNAc)6, GlcNAc, and glucosamine reveal a unique binding mode suitable for interaction with de-N-acetylated PNAG (dPNAG). Furthermore, PgaB310-672 contains a β-hairpin loop (βHL) important for binding PNAG that was disordered in previous PgaB42-655 structures and is highly dynamic in the MD simulations. We propose that conformational changes in PgaB310-672 mediated by the βHL on binding of PNAG/dPNAG play an important role in the targeting of the polymer for export and its release.
Organizational Affiliation:
Program in Molecular Structure and Function, Research Institute, The Hospital for Sick Children, Toronto, ON, Canada M5G 1X8;Department of Biochemistry, University of Toronto, Toronto, ON, Canada M5S 1A8;