Acoustic transfer of protein crystals from agarose pedestals to micromeshes for high-throughput screening.
Cuttitta, C.M., Ericson, D.L., Scalia, A., Roessler, C.G., Teplitsky, E., Joshi, K., Campos, O., Agarwal, R., Allaire, M., Orville, A.M., Sweet, R.M., Soares, A.S.(2015) Acta Crystallogr D Biol Crystallogr 71: 94-103
- PubMed: 25615864
- DOI: https://doi.org/10.1107/S1399004714013728
- Primary Citation of Related Structures:
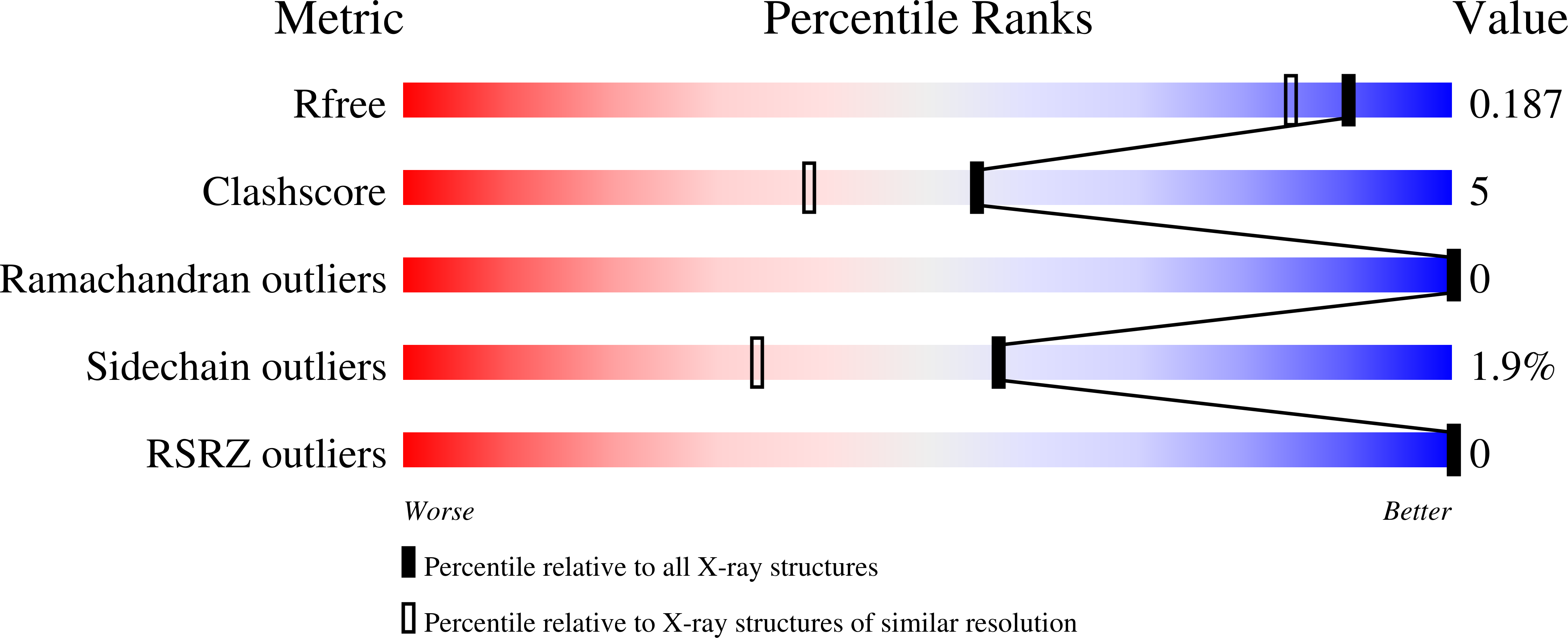
4P2E - PubMed Abstract:
Acoustic droplet ejection (ADE) is an emerging technology with broad applications in serial crystallography such as growing, improving and manipulating protein crystals. One application of this technology is to gently transfer crystals onto MiTeGen micromeshes with minimal solvent. Once mounted on a micromesh, each crystal can be combined with different chemicals such as crystal-improving additives or a fragment library. Acoustic crystal mounting is fast (2.33 transfers s(-1)) and all transfers occur in a sealed environment that is in vapor equilibrium with the mother liquor. Here, a system is presented to retain crystals near the ejection point and away from the inaccessible dead volume at the bottom of the well by placing the crystals on a concave agarose pedestal (CAP) with the same chemical composition as the crystal mother liquor. The bowl-shaped CAP is impenetrable to crystals. Consequently, gravity will gently move the crystals into the optimal location for acoustic ejection. It is demonstrated that an agarose pedestal of this type is compatible with most commercially available crystallization conditions and that protein crystals are readily transferred from the agarose pedestal onto micromeshes with no loss in diffraction quality. It is also shown that crystals can be grown directly on CAPs, which avoids the need to transfer the crystals from the hanging drop to a CAP. This technology has been used to combine thermolysin and lysozyme crystals with an assortment of anomalously scattering heavy atoms. The results point towards a fast nanolitre method for crystal mounting and high-throughput screening.
Organizational Affiliation:
Office of Educational Programs, Brookhaven National Laboratory, Upton, NY 11973-5000, USA.