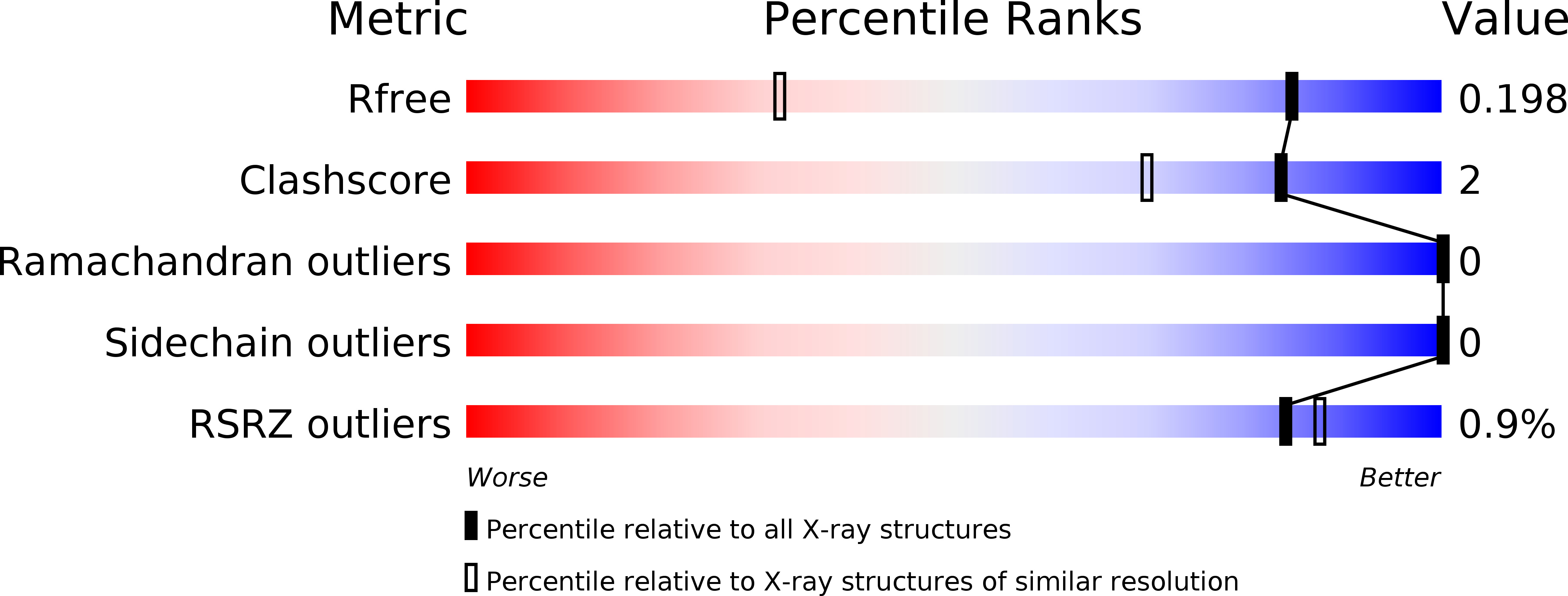
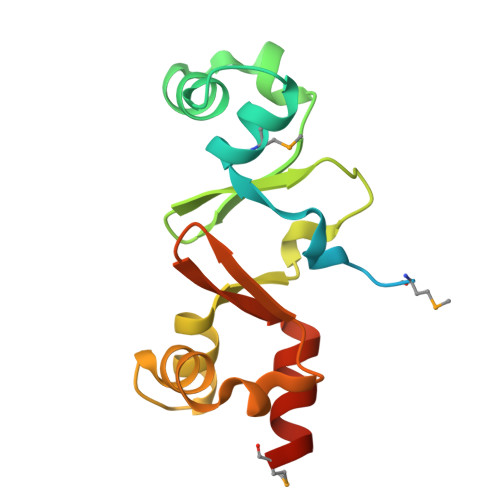
Structure of the double-stranded DNA-binding type IV secretion protein TraN from Enterococcus.
Goessweiner-Mohr, N., Eder, M., Hofer, G., Fercher, C., Arends, K., Birner-Gruenberger, R., Grohmann, E., Keller, W.(2014) Acta Crystallogr D Biol Crystallogr 70: 2376-2389
- PubMed: 25195751
- DOI: https://doi.org/10.1107/S1399004714014187
- Primary Citation of Related Structures:
4P0Y, 4P0Z, 4PM3 - PubMed Abstract:
Conjugative transfer through type IV secretion multiprotein complexes is the most important means of spreading antimicrobial resistance. Plasmid pIP501, frequently found in clinical Enterococcus faecalis and Enterococcus faecium isolates, is the first Gram-positive (G+) conjugative plasmid for which self-transfer to Gram-negative (G-) bacteria has been demonstrated. The pIP501-encoded type IV secretion system (T4SS) protein TraN localizes to the cytoplasm and shows specific DNA binding. The specific DNA-binding site upstream of the pIP501 origin of transfer (oriT) was identified by a novel footprinting technique based on exonuclease digestion and sequencing, suggesting TraN to be an accessory protein of the pIP501 relaxase TraA. The structure of TraN was determined to 1.35 Å resolution. It revealed an internal dimer fold with antiparallel β-sheets in the centre and a helix-turn-helix (HTH) motif at both ends. Surprisingly, structurally related proteins (excisionases from T4SSs of G+ conjugative transposons and transcriptional regulators of the MerR family) resembling only one half of TraN were found. Thus, TraN may be involved in the early steps of pIP501 transfer, possibly triggering pIP501 TraA relaxase activity by recruiting the relaxosome to the assembled mating pore.
Organizational Affiliation:
Institute of Molecular Biosciences, University of Graz, Humboldtstrasse 50/III, 8010 Graz, Austria.