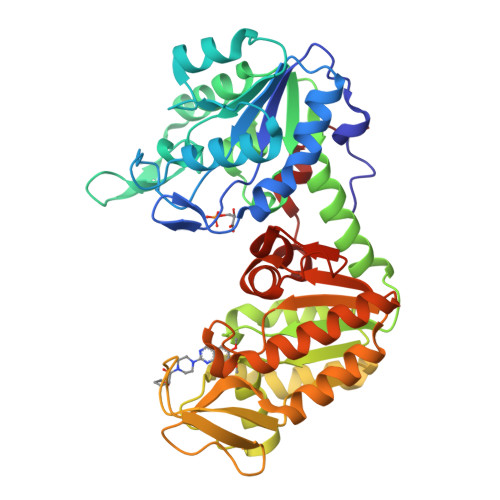
Terazosin activates Pgk1 and Hsp90 to promote stress resistance.
Chen, X., Zhao, C., Li, X., Wang, T., Li, Y., Cao, C., Ding, Y., Dong, M., Finci, L., Wang, J.H., Li, X., Liu, L.(2015) Nat Chem Biol 11: 19-25
- PubMed: 25383758
- DOI: https://doi.org/10.1038/nchembio.1657
- Primary Citation of Related Structures:
4O33, 4O3F - PubMed Abstract:
Drugs that can protect against organ damage are urgently needed, especially for diseases such as sepsis and brain stroke. We discovered that terazosin (TZ), a widely marketed α1-adrenergic receptor antagonist, alleviated organ damage and improved survival in rodent models of stroke and sepsis. Through combined studies of enzymology and X-ray crystallography, we discovered that TZ binds a new target, phosphoglycerate kinase 1 (Pgk1), and activates its enzymatic activity, probably through 2,4-diamino-6,7-dimethoxyisoquinoline's ability to promote ATP release from Pgk1. Mechanistically, the ATP generated from Pgk1 may enhance the chaperone activity of Hsp90, an ATPase known to associate with Pgk1. Upon activation, Hsp90 promotes multistress resistance. Our studies demonstrate that TZ has a new protein target, Pgk1, and reveal its corresponding biological effect. As a clinical drug, TZ may be quickly translated into treatments for diseases including stroke and sepsis.
Organizational Affiliation:
1] State Key Laboratory of Biomembrane and Membrane Biotechnology, School of Life Sciences, Peking University, Beijing, China. [2] Beijing Institute for Brain Disorder and Beijing Tiantan Hospital, Capital Medical University, Beijing, China.