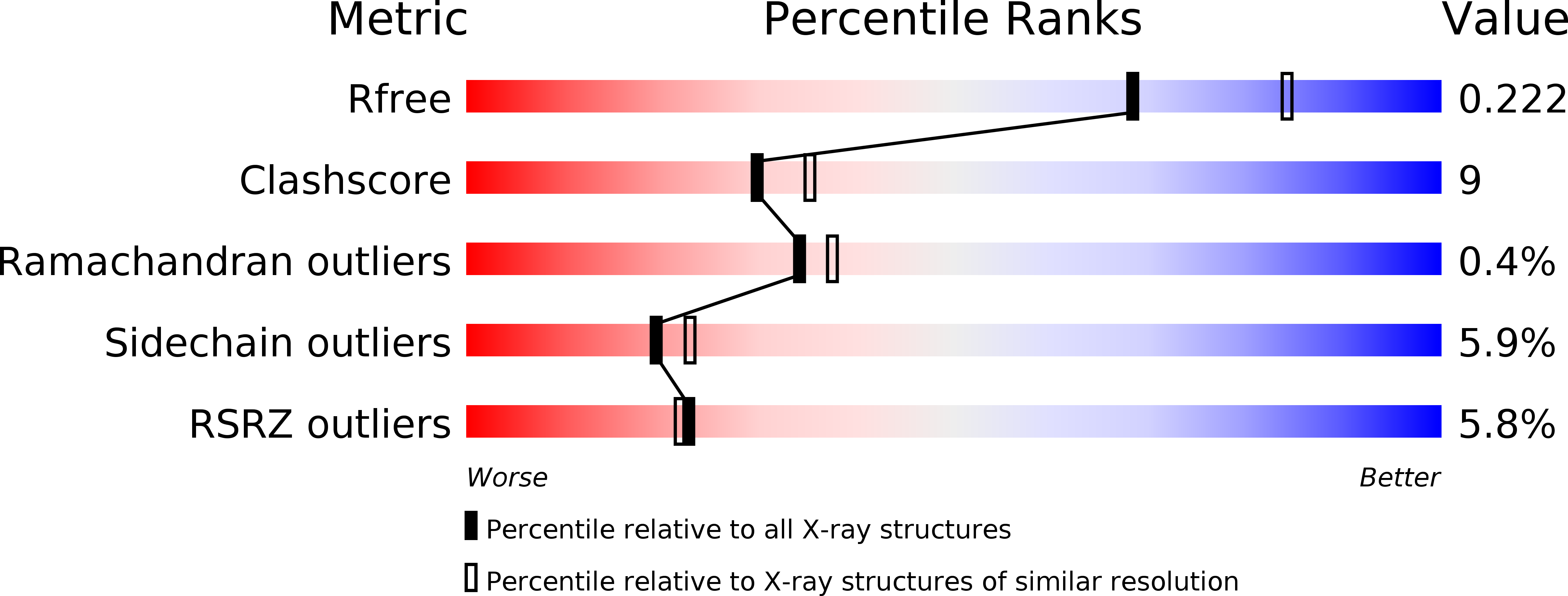
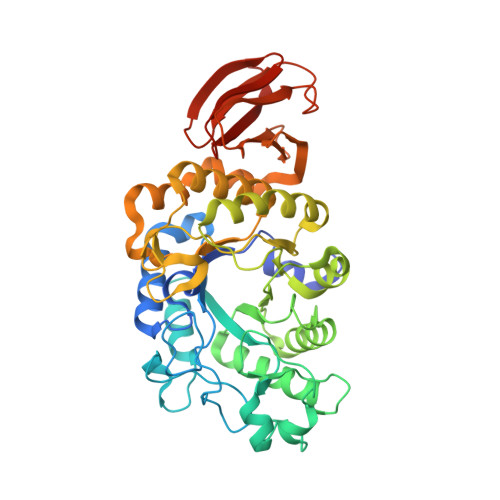
Structure-specificity relationships in Abp, a GH27 beta-L-arabinopyranosidase from Geobacillus stearothermophilus T6
Lansky, S., Salama, R., Solomon, H.V., Feinberg, H., Belrhali, H., Shoham, Y., Shoham, G.(2014) Acta Crystallogr D Biol Crystallogr 70: 2994-3012
- PubMed: 25372689
- DOI: https://doi.org/10.1107/S139900471401863X
- Primary Citation of Related Structures:
4NX0, 4NXK, 4NZF - PubMed Abstract:
L-Arabinose sugar residues are relatively abundant in plants and are found mainly in arabinan polysaccharides and in other arabinose-containing polysaccharides such as arabinoxylans and pectic arabinogalactans. The majority of the arabinose units in plants are present in the furanose form and only a small fraction of them are present in the pyranose form. The L-arabinan-utilization system in Geobacillus stearothermophilus T6, a Gram-positive thermophilic soil bacterium, has recently been characterized, and one of the key enzymes was found to be an intracellular β-L-arabinopyranosidase (Abp). Abp, a GH27 enzyme, was shown to remove β-L-arabinopyranose residues from synthetic substrates and from the native substrates sugar beet arabinan and larch arabinogalactan. The Abp monomer is made up of 448 amino acids, and based on sequence homology it was suggested that Asp197 is the catalytic nucleophile and Asp255 is the catalytic acid/base. In the current study, the detailed three-dimensional structure of wild-type Abp (at 2.28 Å resolution) and its catalytic mutant Abp-D197A with (at 2.20 Å resolution) and without (at 2.30 Å resolution) a bound L-arabinose product are reported as determined by X-ray crystallography. These structures demonstrate that the three-dimensional structure of the Abp monomer correlates with the general fold observed for GH27 proteins, consisting of two main domains: an N-terminal TIM-barrel domain and a C-terminal all-β domain. The two catalytic residues are located in the TIM-barrel domain, such that their carboxylic functional groups are about 5.9 Å from each other, consistent with a retaining mechanism. An isoleucine residue (Ile67) located at a key position in the active site is shown to play a critical role in the substrate specificity of Abp, providing a structural basis for the high preference of the enzyme towards arabinopyranoside over galactopyranoside substrates. The crystal structure demonstrates that Abp is a tetramer made up of two `open-pincers' dimers, which clamp around each other to form a central cavity. The four active sites of the Abp tetramer are situated on the inner surface of this cavity, all opening into the central space of the cavity. The biological relevance of this tetrameric structure is supported by independent results obtained from size-exclusion chromatography (SEC), dynamic light-scattering (DLS) and small-angle X-ray scattering (SAXS) experiments. These data and their comparison to the structural data of related GH27 enzymes are used for a more general discussion concerning structure-selectivity aspects in this glycoside hydrolase (GH) family.
Organizational Affiliation:
Institute of Chemistry and the Laboratory for Structural Chemistry and Biology, The Hebrew University of Jerusalem, Jerusalem 91904, Israel.