Red Blood Cell Invasion by Plasmodium vivax: Structural Basis for DBP Engagement of DARC.
Batchelor, J.D., Malpede, B.M., Omattage, N.S., Dekoster, G.T., Henzler-Wildman, K.A., Tolia, N.H.(2014) PLoS Pathog 10: e1003869-e1003869
- PubMed: 24415938
- DOI: https://doi.org/10.1371/journal.ppat.1003869
- Primary Citation of Related Structures:
4NUU, 4NUV - PubMed Abstract:
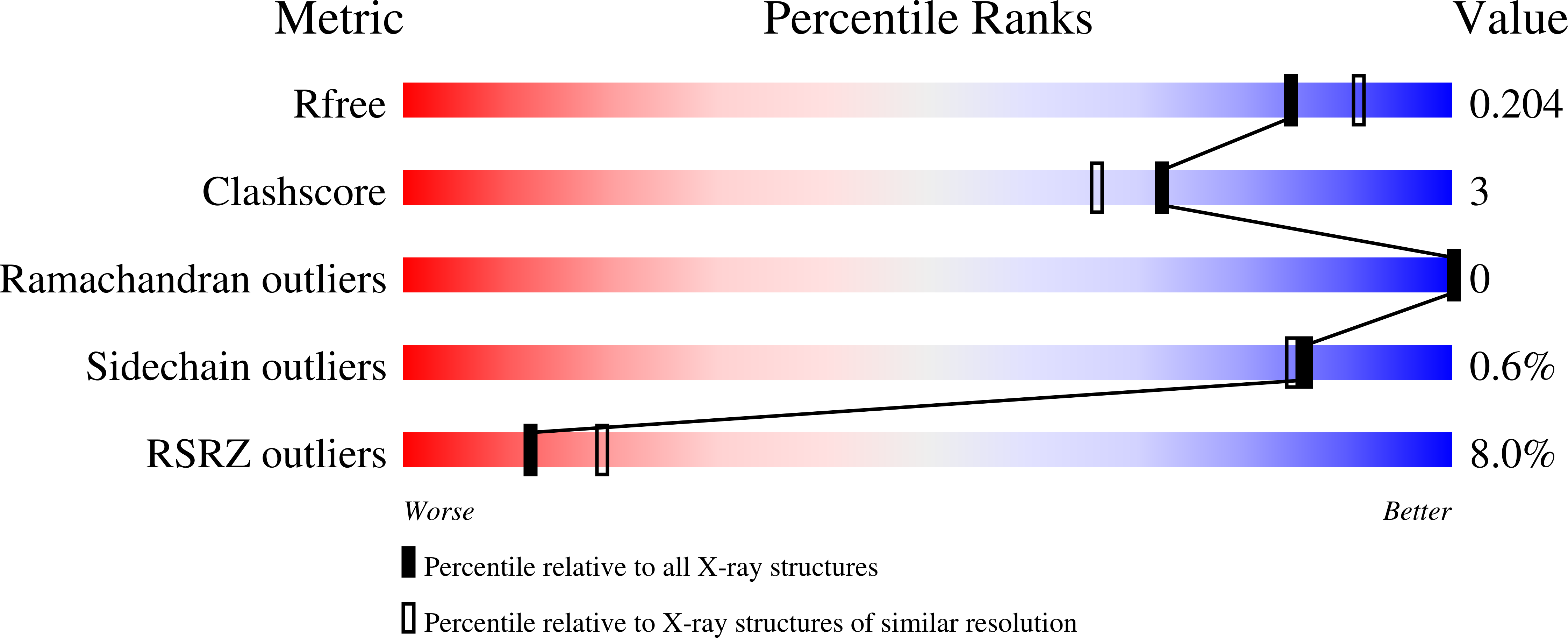
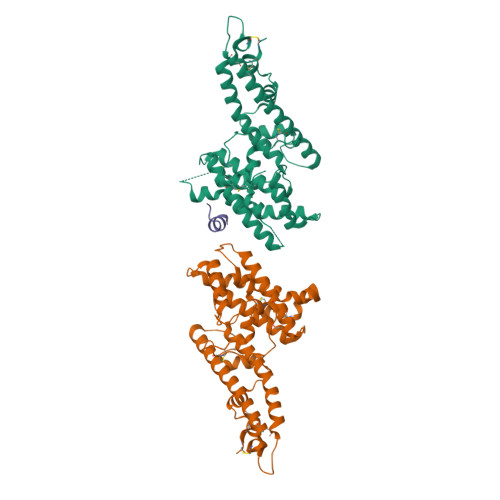
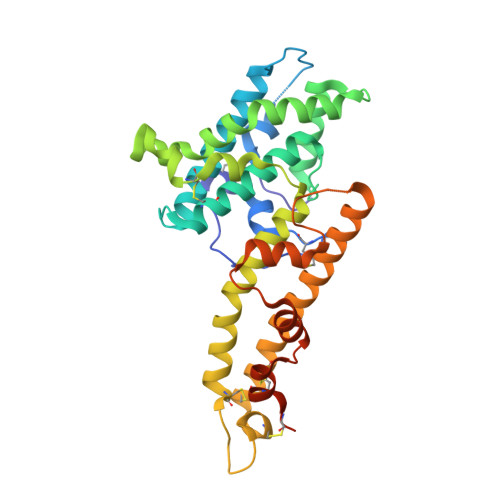
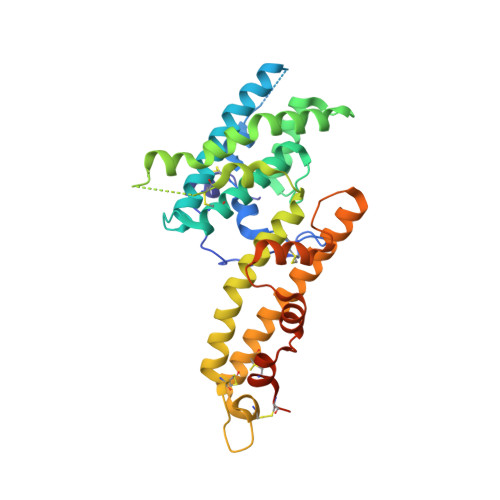
Plasmodium parasites use specialized ligands which bind to red blood cell (RBC) receptors during invasion. Defining the mechanism of receptor recognition is essential for the design of interventions against malaria. Here, we present the structural basis for Duffy antigen (DARC) engagement by P. vivax Duffy binding protein (DBP). We used NMR to map the core region of the DARC ectodomain contacted by the receptor binding domain of DBP (DBP-RII) and solved two distinct crystal structures of DBP-RII bound to this core region of DARC. Isothermal titration calorimetry studies show these structures are part of a multi-step binding pathway, and individual point mutations of residues contacting DARC result in a complete loss of RBC binding by DBP-RII. Two DBP-RII molecules sandwich either one or two DARC ectodomains, creating distinct heterotrimeric and heterotetrameric architectures. The DARC N-terminus forms an amphipathic helix upon DBP-RII binding. The studies reveal a receptor binding pocket in DBP and critical contacts in DARC, reveal novel targets for intervention, and suggest that targeting the critical DARC binding sites will lead to potent disruption of RBC engagement as complex assembly is dependent on DARC binding. These results allow for models to examine inter-species infection barriers, Plasmodium immune evasion mechanisms, P. knowlesi receptor-ligand specificity, and mechanisms of naturally acquired P. vivax immunity. The step-wise binding model identifies a possible mechanism by which signaling pathways could be activated during invasion. It is anticipated that the structural basis of DBP host-cell engagement will enable development of rational therapeutics targeting this interaction.
Organizational Affiliation:
Department of Molecular Microbiology and Microbial Pathogenesis, Washington University School of Medicine, Saint Louis, Missouri, United States of America.