Molecular mechanism for self-protection against the type VI secretion system in Vibrio cholerae.
Yang, X., Xu, M., Wang, Y., Xia, P., Wang, S., Ye, B., Tong, L., Jiang, T., Fan, Z.(2014) Acta Crystallogr D Biol Crystallogr 70: 1094-1103
- PubMed: 24699653
- DOI: https://doi.org/10.1107/S1399004714001242
- Primary Citation of Related Structures:
4NOO - PubMed Abstract:
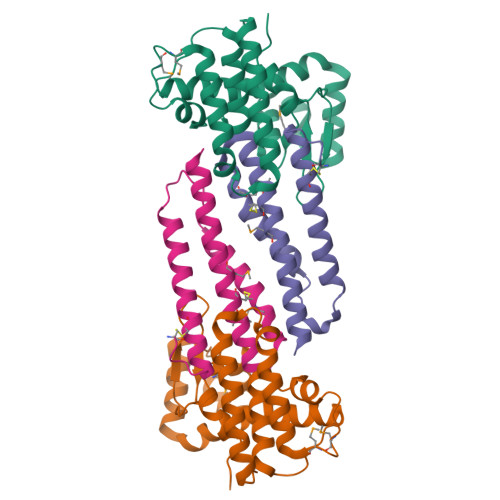
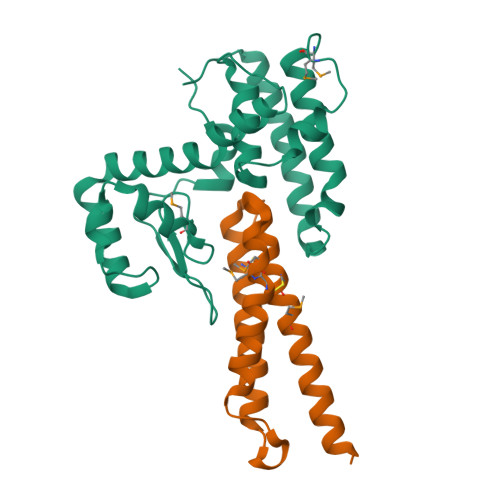
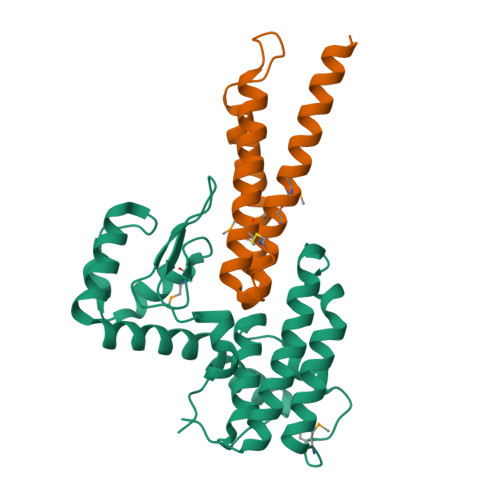
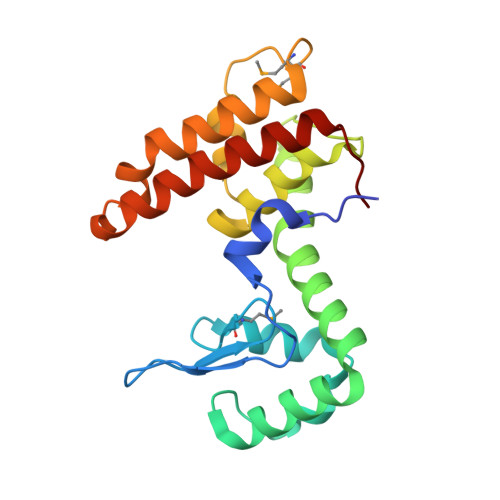
VgrG proteins form the spike of the type VI secretion system (T6SS) syringe-like complex. VgrG3 of Vibrio cholerae degrades the peptidoglycan cell wall of rival bacteria via its C-terminal region (VgrG3C) through its muramidase activity. VgrG3C consists of a peptidoglycan-binding domain (VgrG3C(PGB)) and a putative catalytic domain (VgrG3C(CD)), and its activity can be inhibited by its immunity protein partner TsiV3. Here, the crystal structure of V. cholerae VgrG3C(CD) in complex with TsiV3 is presented at 2.3 Å resolution. VgrG3C(CD) adopts a chitosanase fold. A dimer of TsiV3 is bound in the deep active-site groove of VgrG3C(CD), occluding substrate binding and distorting the conformation of the catalytic dyad. Gln91 and Arg92 of TsiV3 are located in the centre of the interface and are important for recognition of VgrG3C. Mutation of these residues destabilized the complex and abolished the inhibitory activity of TsiV3 against VgrG3C toxicity in cells. Disruption of TsiV3 dimerization also weakened the complex and impaired the inhibitory activity. These structural, biochemical and functional data define the molecular mechanism underlying the self-protection of V. cholerae and expand the understanding of the role of T6SS in bacterial competition.
Organizational Affiliation:
Chinese Academy of Sciences Key Laboratory of Infection and Immunity, Institute of Biophysics, Chinese Academy of Sciences, Beijing 100101, People's Republic of China.