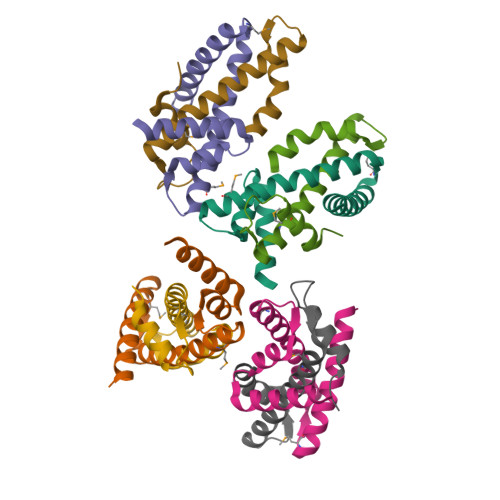
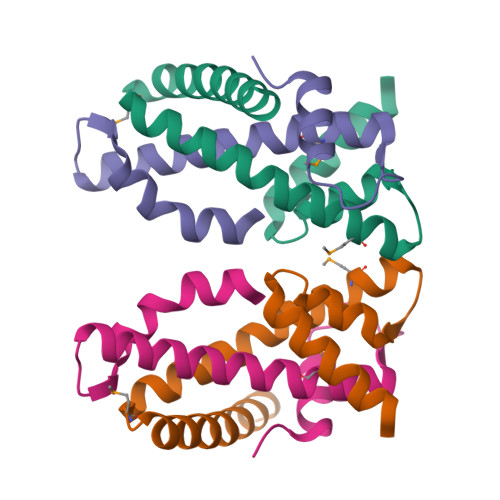
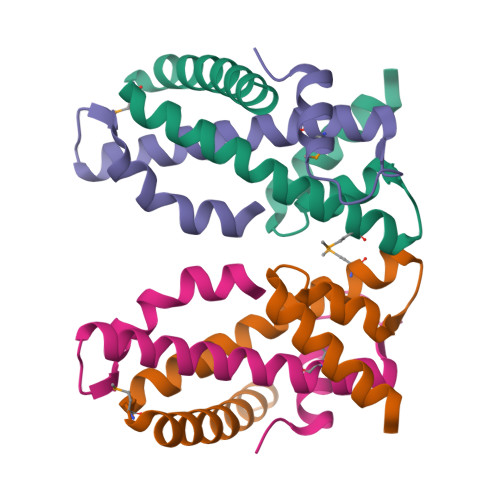
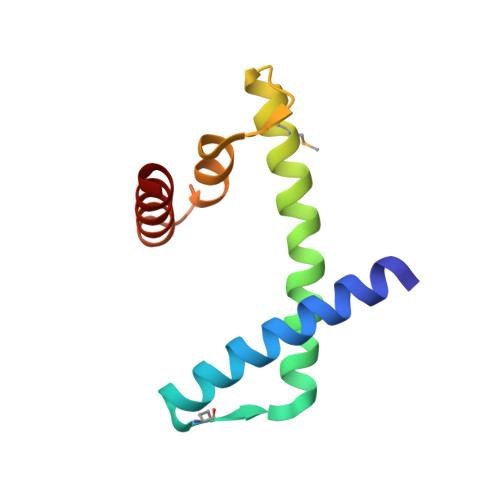
The MHF complex senses branched DNA by binding a pair of crossover DNA duplexes.
Zhao, Q., Saro, D., Sachpatzidis, A., Singh, T.R., Schlingman, D., Zheng, X.F., Mack, A., Tsai, M.S., Mochrie, S., Regan, L., Meetei, A.R., Sung, P., Xiong, Y.(2014) Nat Commun 5: 2987-2987
- PubMed: 24390579
- DOI: https://doi.org/10.1038/ncomms3987
- Primary Citation of Related Structures:
4NDY, 4NE1, 4NE3, 4NE5, 4NE6 - PubMed Abstract:
The conserved MHF1-MHF2 (MHF) complex functions in the activation of the Fanconi anaemia pathway of the DNA damage response, in regulating homologous recombination, and in DNA replication fork maintenance. MHF facilitates the processing of multiple types of branched DNAs by the DNA translocase FANCM. Here we report the crystal structure of a human MHF-DNA complex that reveals the DNA-binding mode of MHF. The structure suggests that MHF prefers branched DNA over double-stranded DNA because it engages two duplex arms. Biochemical analyses verify that MHF preferentially engages DNA forks or various four-way junctions independent of the junction-site structure. Furthermore, genetic experiments provide evidence that the observed DNA-binding interface of MHF is important for cellular resistance to DNA damage. These results offer insights into how the MHF complex recognizes branched DNA and stimulates FANCM activity at such a structure to promote genome maintenance.
Organizational Affiliation:
1] Department of Molecular Biophysics and Biochemistry, Yale University School of Medicine, New Haven, Connecticut 06520, USA [2].