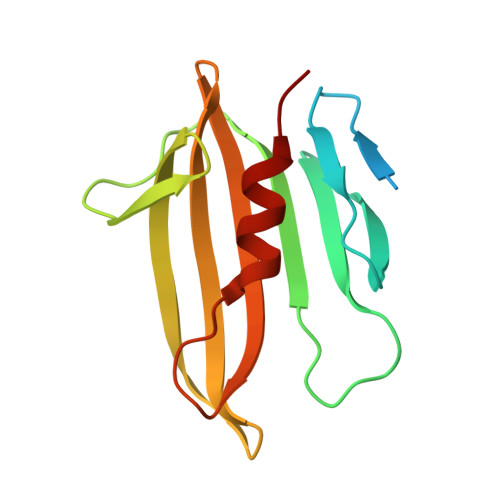
Structural insights into the T6SS effector protein Tse3 and the Tse3-Tsi3 complex from Pseudomonas aeruginosa reveal a calcium-dependent membrane-binding mechanism
Lu, D., Shang, G., Zhang, H., Yu, Q., Cong, X., Yuan, J., He, F., Zhu, C., Zhao, Y., Yin, K., Chen, Y., Hu, J., Zhang, X., Yuan, Z., Xu, S., Hu, W., Cang, H., Gu, L.(2014) Mol Microbiol 92: 1092-1112
- PubMed: 24724564
- DOI: https://doi.org/10.1111/mmi.12616
- Primary Citation of Related Structures:
4M5E, 4M5F, 4N7S, 4N80, 4N88 - PubMed Abstract:
The opportunistic pathogen Pseudomonas aeruginosa uses the type VI secretion system (T6SS) to deliver the muramidase Tse3 into the periplasm of rival bacteria to degrade their peptidoglycan (PG). Concomitantly, P. aeruginosa uses the periplasm-localized immunity protein Tsi3 to prevent potential self-intoxication caused by Tse3, and thus gains an edge over rival bacteria in fierce niche competition. Here, we report the crystal structures of Tse3 and the Tse3-Tsi3 complex. Tse3 contains an annexin repeat-like fold at the N-terminus and a G-type lysozyme fold at the C-terminus. One loop in the N-terminal domain (Loop 12) and one helix (α9) from the C-terminal domain together anchor Tse3 and the Tse3-Tsi3 complex to membrane in a calcium-dependent manner in vitro, and this membrane-binding ability is essential for Tse3's activity. In the C-terminal domain, a Y-shaped groove present on the surface likely serves as the PG binding site. Two calcium-binding motifs are also observed in the groove and these are necessary for Tse3 activity. In the Tse3-Tsi3 structure, three loops of Tsi3 insert into the substrate-binding groove of Tse3, and three calcium ions present at the interface of the complex are indispensable for the formation of the Tse3-Tsi3 complex.
Organizational Affiliation:
State Key Laboratory of Microbial Technology, Shandong University, Jinan, 250100, Shandong, China; The Liver Centre of Fujian Province, MengChao Hepatobiliary Hospital of Fujian Medical University, Fuzhou, 350025, Fujian, China.